MET Target Analysis Report Summary


About the Target
c-Met (also referred to as MET) plays a crucial role in various biological processes and is associated with different diseases. In hepatoblastoma (HB), mutant beta-catenin escapes degradation and enters the nucleus, activating the transcription of target genes [1]. Additionally, HB tumor cells can suppress beta-catenin degradation through CAPRIN2-activating mutations, leading to beta-catenin accumulation [1]. Notably, c-Met activation by hepatocyte growth factor (HGF) induces beta-catenin translocation to the nucleus via tyrosine phosphorylation [1]. Studies have shown a positive correlation between activated c-Met and nuclear accumulation of beta-catenin in human HB specimens [1].
In melanoma, c-MET can be upregulated through transcriptional regulation by MITF and reduced levels of miRNAs targeting c-MET transcript [2]. Elevated levels of HGF, produced by both stromal cells and melanoma cells, enhance c-MET activity, contributing to melanoma progression [2]. Autocrine HGF/c-MET signaling supports melanoma cell proliferation, survival, motility, invasiveness, and niche formation [2].
In a study utilizing JNJ-61186372 BsAb, it was found that high EGFR to c-MET expression confers increased potency in blocking HGF-mediated phosphorylation, highlighting the role of c-MET in target engagement [3].
MET amplification has been associated with EGFR-TKI resistance in certain cancers [4]. It activates EGFR-independent phosphorylation of ErbB3, leading to downstream activation of the PI3K/AKT pathway, which bypasses the effects of EGFR-TKI [4].
Lnc-TALC, a long non-coding RNA (lncRNA), exerts its effect on glioblastoma (GBM) by regulating the c-Met signaling pathway as a competitive RNA [5]. The mechanistic scheme of lnc-TALC involves promoting MGMT expression, potentially serving as a therapeutic target to overcome resistance to TMZ chemotherapy in GBM patients [5].
Overall, c-Met (MET) has been implicated in various disease contexts, modulating biological processes, drug resistance, and potential therapeutic targets.
Based on the provided context information, the key viewpoints regarding c-MET (MET) are as follows:
Targeting the HGF/c-MET interaction could be effective in countering microenvironment-driven support to leukemic cells. Combinatorial therapies utilizing conventional drugs along with c-MET kinase inhibitors or exosomes-delivered effector molecules could enhance therapeutic approaches [6].
Loss of ABHD5 releases DPY30 to translocate into the nucleus, leading to increased YAP methylation and activation. This promotes the transcription of c-Met, contributing to the development and progression of colorectal cancers [7].
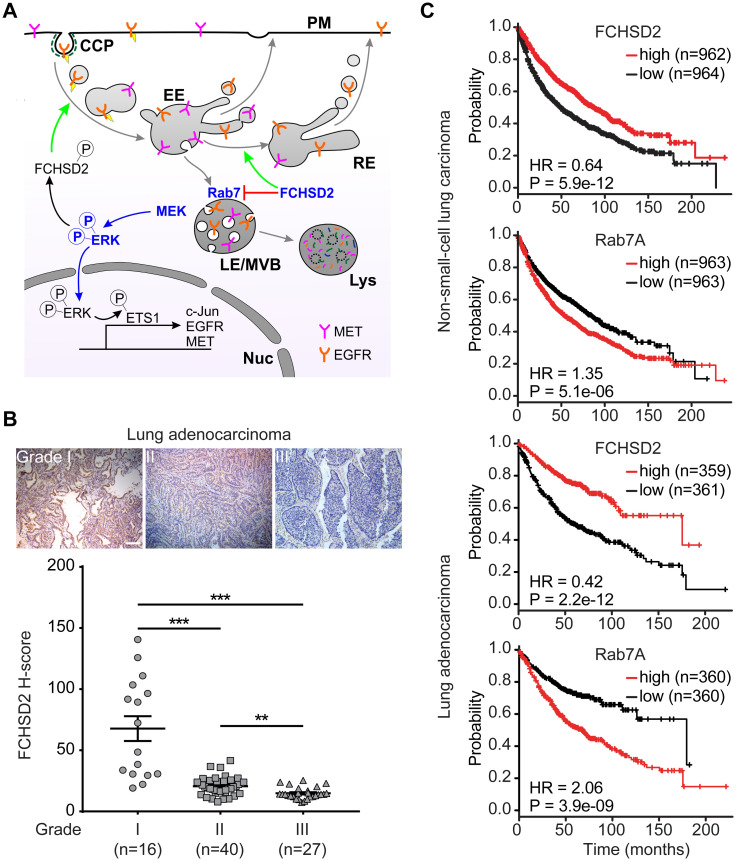
FCHSD2 regulates multiple steps in endocytic trafficking and alters downstream signaling of receptor tyrosine kinases (RTKs). Loss of FCHSD2 leads to the accumulation of RTKs in late endosomes/lysosomes and enhances the transcription and expression of c-Jun, EGFR, and c-Met. The expression of FCHSD2 is decreased in higher grades of lung adenocarcinoma tumors, and patients with higher FCHSD2 expression have better survival rates [8].
In summary, targeting the HGF/c-MET interaction and the regulation of c-MET expression through ABHD5 and FCHSD2 have potential implications for therapeutic interventions in CLL and colorectal and lung cancers, respectively [6][7][8].
Figure [1]
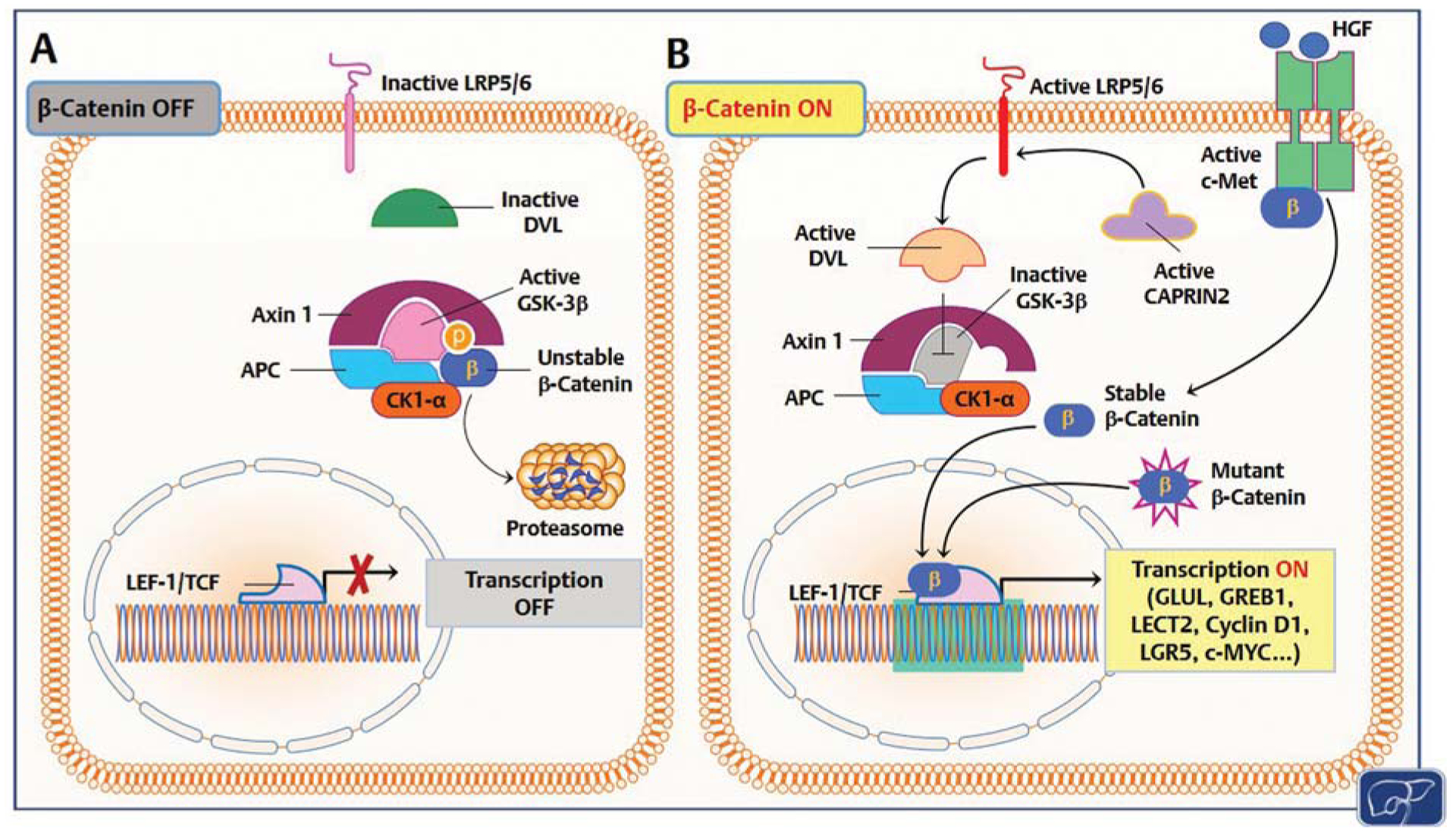
Figure [2]
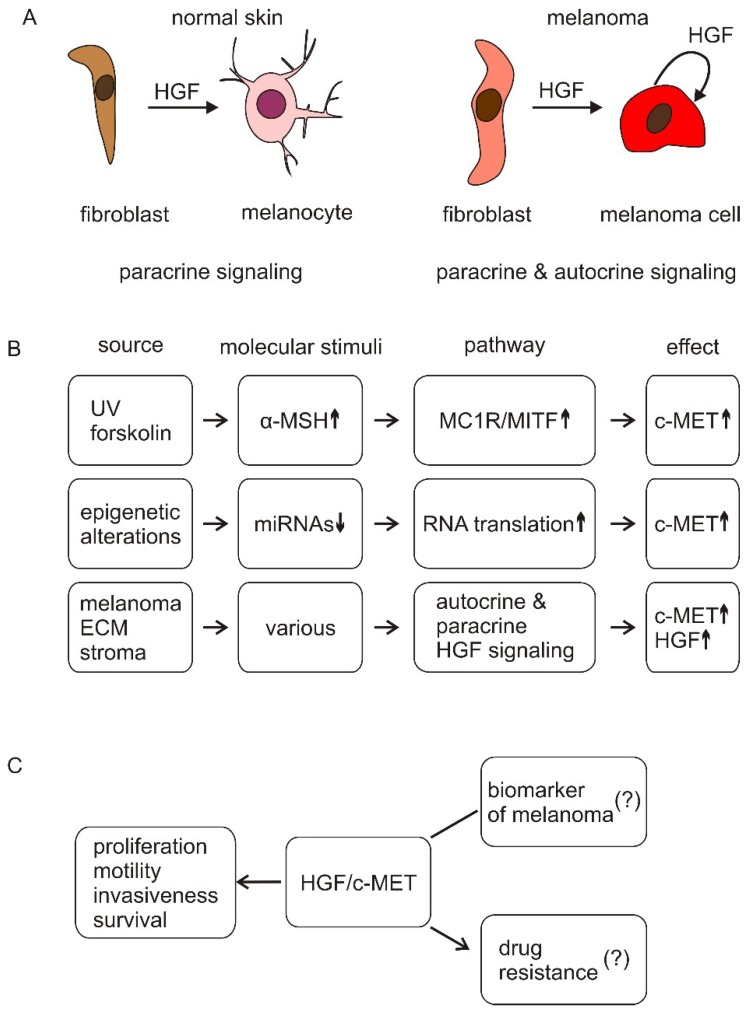
Figure [3]
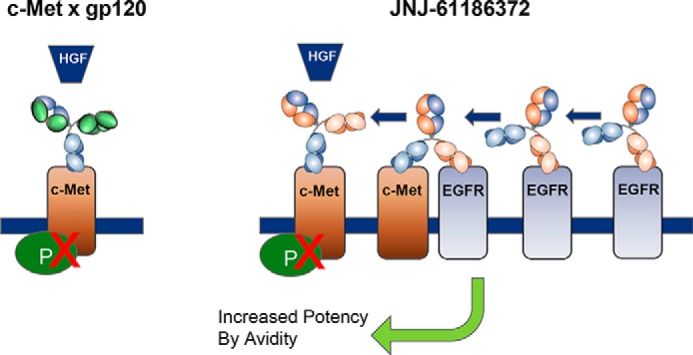
Figure [4]
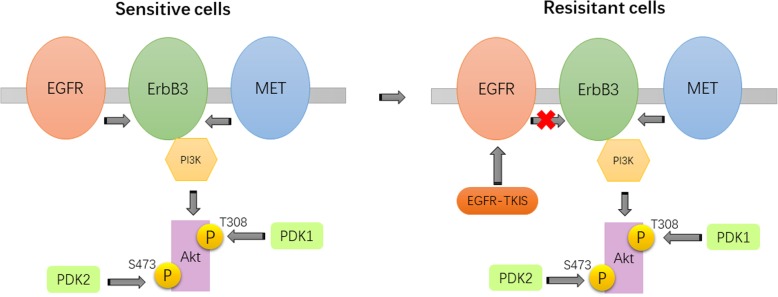
Figure [5]
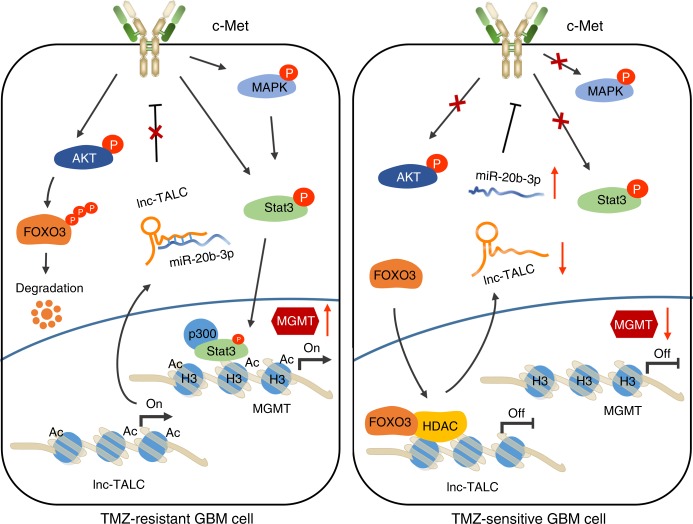
Figure [6]

Figure [7]
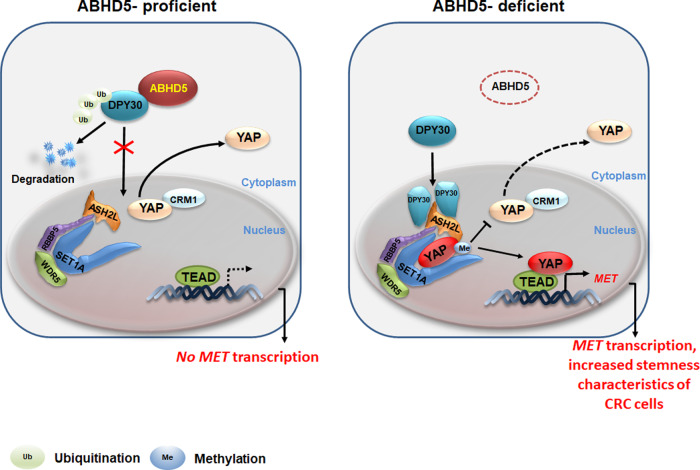
Figure [8]
Note: If you are interested in the full version of this target analysis report, or if you'd like to learn how our AI-powered BDE-Chem can design therapeutic molecules to interact with the MET target at a cost 90% lower than traditional approaches, please feel free to contact us at BD@silexon.ai.
More Common Targets
ABCB1 | ABCG2 | ACE2 | AHR | AKT1 | ALK | AR | ATM | BAX | BCL2 | BCL2L1 | BECN1 | BRAF | BRCA1 | CAMP | CASP3 | CASP9 | CCL5 | CCND1 | CD274 | CD4 | CD8A | CDH1 | CDKN1A | CDKN2A | CREB1 | CXCL8 | CXCR4 | DNMT1 | EGF | EGFR | EP300 | ERBB2 | EREG | ESR1 | EZH2 | FN1 | FOXO3 | HDAC9 | HGF | HMGB1 | HSP90AA1 | HSPA4 | HSPA5 | IDO1 | IFNA1 | IGF1 | IGF1R | IL17A | IL6 | INS | JUN | KRAS | MAPK1 | MAPK14 | MAPK3 | MAPK8 | MAPT | MCL1 | MDM2 | MET | MMP9 | MTOR | MYC | NFE2L2 | NLRP3 | NOTCH1 | PARP1 | PCNA | PDCD1 | PLK1 | PRKAA1 | PRKAA2 | PTEN | PTGS2 | PTK2 | RELA | SIRT1 | SLTM | SMAD4 | SOD1 | SQSTM1 | SRC | STAT1 | STAT3 | STAT5A | TAK1 | TERT | TLR4 | TNF | TP53 | TXN | VEGFA | YAP1

