ESR1 Target Analysis Report Summary


About the Target
Based on the provided context information, several viewpoints about the estrogen receptor (ER/ESR1) can be extracted:
ERα, encoded by the ESR1 gene, contains different structural domains that determine its transcriptional and epigenetic activities, including the N-terminal AF1, hinge domain, AF2 within the C-terminal LBD, and DBD [1].
Activated ERα can recruit coactivators or corepressors to mediate gene transcription or repression, respectively. Coactivators include members of the p160 family, P300/CBP, SWI/SNF complex, PRMTs, and the Mediator complex, while corepressors include NCoR1, NCoR2, and LCoR [1].
There are differences in ER's coregulatory proteins and genomic binding sites between endometrial cancer and breast cancer cells. Pioneer factors such as FOXA1 and GATA3 play a key role in enabling ER genomic binding in breast cancer, but their counterparts in endometrial cancer remain unknown. ER cofactors may also differ between the two cancer types [2].
The chromatin landscape and transcription factor repertoire in breast and endometrial cancer cells contribute to different ER binding profiles and target genes. FOXA1 and GATA3 act as pioneer factors for ER in breast cancer cells, but the mechanisms underlying ER's unique genomic binding pattern in endometrial cancer cells are unclear [2].
Functional studies of specific variants of ESR1, such as rs9340803, have been conducted to understand their effects. Decreased ESR1 expression caused by rs9340803 A to G variation may disrupt the balance of cholesterol metabolism, potentially promoting more Abeta production [3].
The classic ER transcriptional mechanism involves the binding of estrogen (E2) to ER, triggering receptor dimerization, nuclear translocation, and exposure of the AF2 site for coactivator binding. ER then recognizes and interacts with estrogen response elements (ERE) in the nucleus, forming a transcriptionally active complex and interacting with coactivators. The p160 family of proteins is an example of ER coactivators [4].
Endocrine therapies target the estrogen signaling pathway through various mechanisms. Aromatase inhibitors block estrogen production, selective estrogen receptor modulators (SERMs) competitively inhibit estrogen binding to ER, and selective estrogen receptor downregulators (SERDs) act as pure ER antagonists. Proteolysis targeting chimeras (PROTACs) induce degradation of ER [5].
These viewpoints provide insights into the structural domains and activities of ER, differences in ER regulation between breast and endometrial cancer, functional studies of specific ESR1 variants, the classic ER transcriptional mechanism, and the mechanisms of action of endocrine therapies.
Based on the given context information, here is a comprehensive summary of the different viewpoints related to the estrogen receptor (also referred to as ER, ESR1) and its molecular targets:
Tocotrienols (a form of vitamin E) have different molecular targets in various cell types, including cancer cells expressing ER, cancer cells not expressing ER, and normal neuronal cells subjected to specific stressors [6].
YB-1 interacts with ERalpha to regulate the stemness and differentiation of ER-positive breast cancer stem cells [7].
In SALL2-hypomethylated ER+ breast cancer, SALL2 activates ER signaling through the upregulation of ESR1, resulting in estrogen-dependent growth and tamoxifen sensitivity. In SALL2-hypermethylated ER+ breast cancer, decreased SALL2 expression represses ERalpha and PTEN expression and activates Akt/mTOR signaling, leading to estrogen-independent tumor growth and tamoxifen resistance. Restoring SALL2 resensitizes tamoxifen-resistant breast cancer to endocrine therapy [8].
The estrogen signaling pathway regulates RASD1 expression in the uterine epithelium. Decreased estrogen levels in patients with recurrent implantation failure (RIF) lead to reduced RASD1 levels in the uterine endometrium, resulting in abnormal implantation [9].
Estrogen and FXR ligands (such as estrogen, bile acids, and gut microbial metabolites) impact ERalpha and FXR signaling in hepatocytes, influencing hepatic lipid metabolism [10].
These viewpoints provide insights into the role of ER and its associated signaling pathways in different contexts, including cancer biology, endometrial function, and hepatic metabolism.
Figure [1]
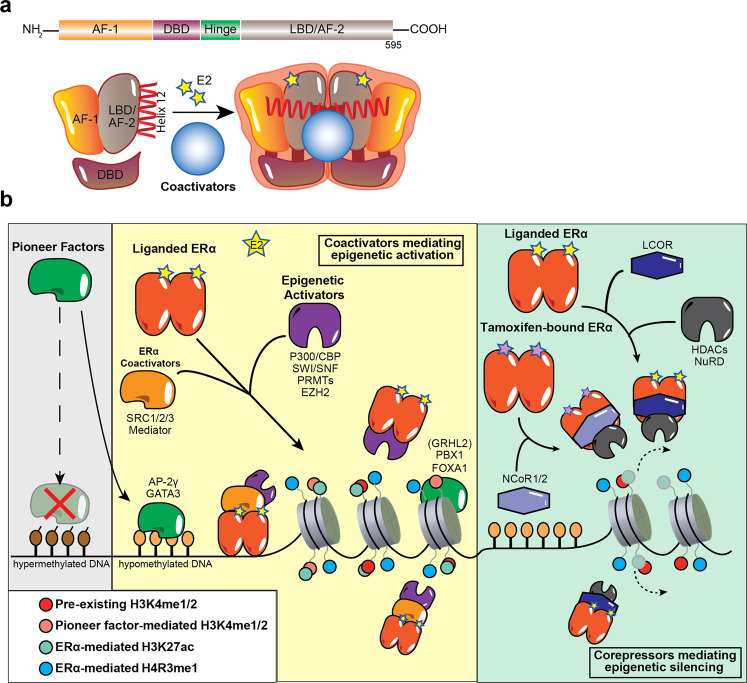
Figure [2]
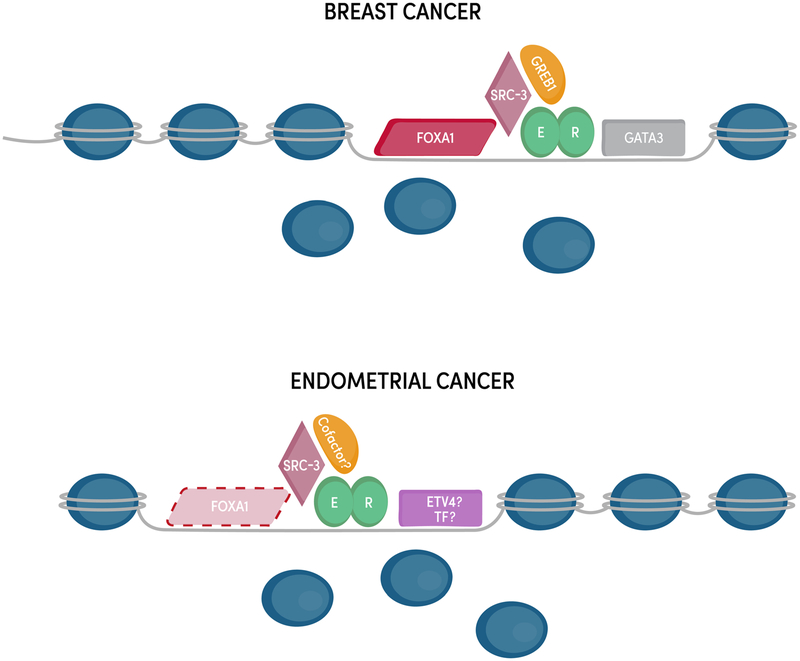
Figure [3]
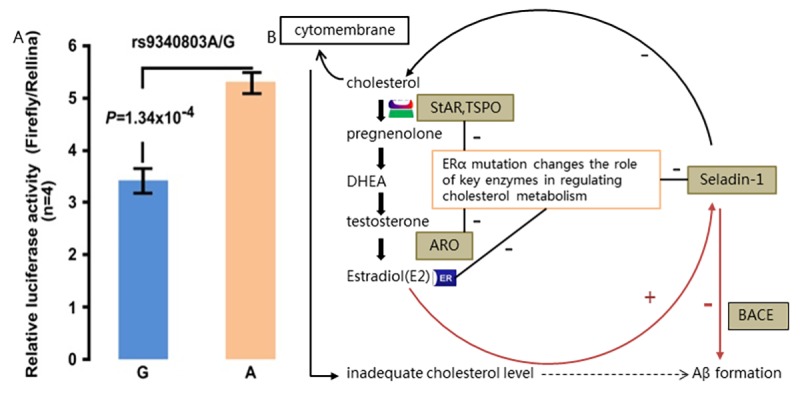
Figure [4]
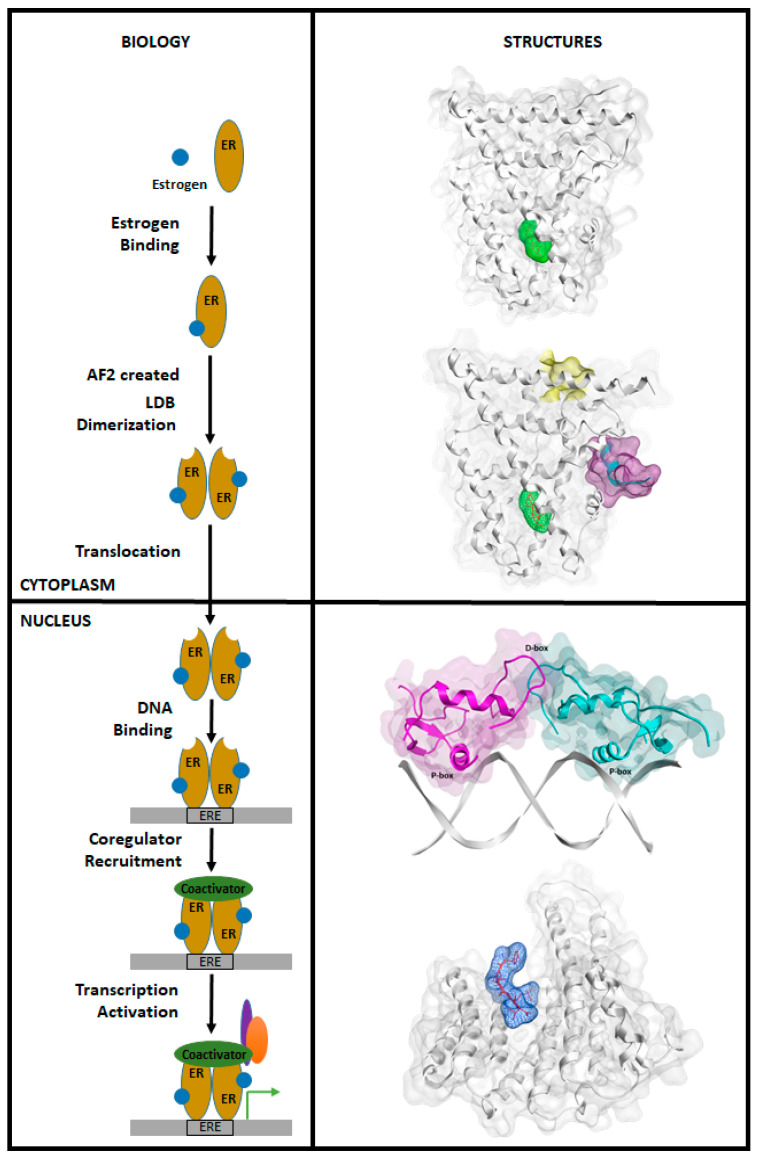
Figure [5]

Figure [6]
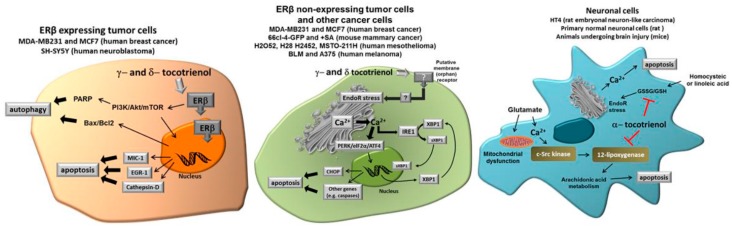
Figure [7]
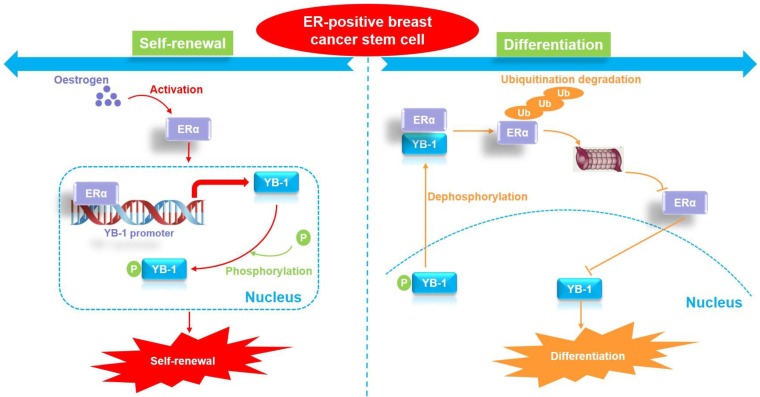
Figure [8]
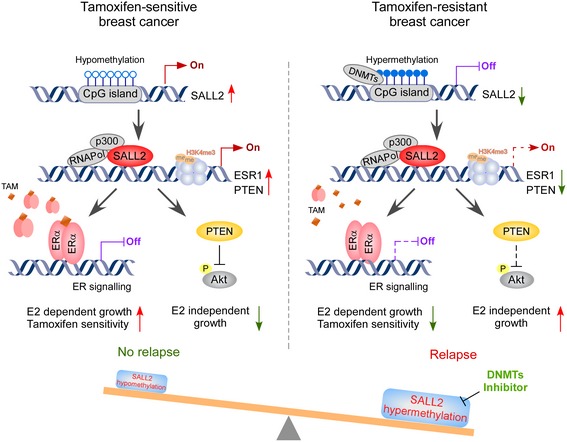
Figure [9]

Figure [10]

Note: If you are interested in the full version of this target analysis report, or if you'd like to learn how our AI-powered BDE-Chem can design therapeutic molecules to interact with the ESR1 target at a cost 90% lower than traditional approaches, please feel free to contact us at BD@silexon.ai.
More Common Targets
ABCB1 | ABCG2 | ACE2 | AHR | AKT1 | ALK | AR | ATM | BAX | BCL2 | BCL2L1 | BECN1 | BRAF | BRCA1 | CAMP | CASP3 | CASP9 | CCL5 | CCND1 | CD274 | CD4 | CD8A | CDH1 | CDKN1A | CDKN2A | CREB1 | CXCL8 | CXCR4 | DNMT1 | EGF | EGFR | EP300 | ERBB2 | EREG | ESR1 | EZH2 | FN1 | FOXO3 | HDAC9 | HGF | HMGB1 | HSP90AA1 | HSPA4 | HSPA5 | IDO1 | IFNA1 | IGF1 | IGF1R | IL17A | IL6 | INS | JUN | KRAS | MAPK1 | MAPK14 | MAPK3 | MAPK8 | MAPT | MCL1 | MDM2 | MET | MMP9 | MTOR | MYC | NFE2L2 | NLRP3 | NOTCH1 | PARP1 | PCNA | PDCD1 | PLK1 | PRKAA1 | PRKAA2 | PTEN | PTGS2 | PTK2 | RELA | SIRT1 | SLTM | SMAD4 | SOD1 | SQSTM1 | SRC | STAT1 | STAT3 | STAT5A | TAK1 | TERT | TLR4 | TNF | TP53 | TXN | VEGFA | YAP1

