NLRP3 Target Analysis Report Summary


About the Target
Based on the provided information, here are some key viewpoints regarding NLRP3:
Glucocorticoid resistance: Patients with idiopathic nephrotic syndrome (INS) may develop resistance to glucocorticoids (GCs) due to epigenetic changes in the NLRP3 gene [1]. This mechanism is related to leukocyte epigenetic changes and may contribute to the development of resistance to GCs in INS patients.
Inflammasome activation: NLRP3 plays a crucial role in both canonical and non-canonical inflammasome activation pathways [2]. Gram-negative bacteria can activate the NLRP3 inflammasome, leading to the release of IL-1beta/IL-18 and pyroptosis. In human monocytes, an alternative NLRP3 inflammasome pathway is activated in response to LPS.
Two-signal model for NLRP3 inflammasome activation: The activation of the NLRP3 inflammasome involves two signals: priming and activation [3]. Priming, which is induced by microbial or endogenous molecules, upregulates NLRP3 expression. Activation occurs through various stimuli like ATP, pore-forming toxins, viral RNA, and particulate matter. K+ efflux, Ca2+ signaling, reactive oxygen species (ROS), mitochondrial dysfunction, and lysosomal rupture are proposed triggers for NLRP3 activation.
Post-translational modifications and regulators of NLRP3: NLRP3 is regulated by post-translational modifications such as phosphorylation, ubiquitination, sumoylation, and S-nitrosylation [4]. These modifications can either positively or negatively affect NLRP3 activation. Additionally, NLRP3 interacts with various proteins and molecules, including ARIH2, BRCC3, FBX12, JNK1, and SENP6/7.
Autophagy and NLRP3: Autophagy plays a role in NLRP3 regulation. Under stress conditions, defective autophagy can lead to the accumulation of damaged mitochondria and activation of the NLRP3 inflammasome, resulting in increased production of IL-1beta and inflammation [5]. On the other hand, p62, an adaptor protein, can stimulate the removal of damaged mitochondria, inhibiting inflammasome activation and pro-inflammatory IL-1beta synthesis.
Overall, NLRP3 is involved in glucocorticoid resistance, inflammasome activation, and various post-translational modifications. Its modulation and regulation are crucial in the context of immune responses and inflammatory processes.
- NLRP3 expression is regulated by various transcription factors, including NF-kappaB, GFI1, AhR, NR1D1, and NFAT5[6].
- NLRP3 inflammasome priming is mediated by post-translational modifications independent of transcription[6].
- Classical activation of the NLRP3 inflammasome involves a two-step model, with signal 1 upregulating NLRP3 expression and signal 2 detecting cellular integrity perturbations[7].
- Alternative inflammasome activation can be triggered by a single signal, such as LPS or an iNKT cell derived signal[7].
- Increased sarcoplasmic reticulum (SR) Ca2+ leak and spontaneous SR Ca2+-release events can lead to NLRP3 inflammasome activation[8].
- Various stimuli, including PAMPs and DAMPs, trigger upstream signaling pathways that converge to activate the NLRP3 inflammasome[9].
- The EPEC pathogen subverts NLRP3 inflammasome assembly through the effector NleA, which inhibits NLRP3 activation and association with ASC[9].
- The NLRP3 inflammasome pathway can be involved in epithelial mesenchymal transition (EMT), potentially promoting the EMT process[10].
Overall, NLRP3 expression and activation are regulated by various transcription factors and signaling pathways, and it plays a role in inflammatory responses, cellular integrity, and EMT processes.
Figure [1]

Figure [2]
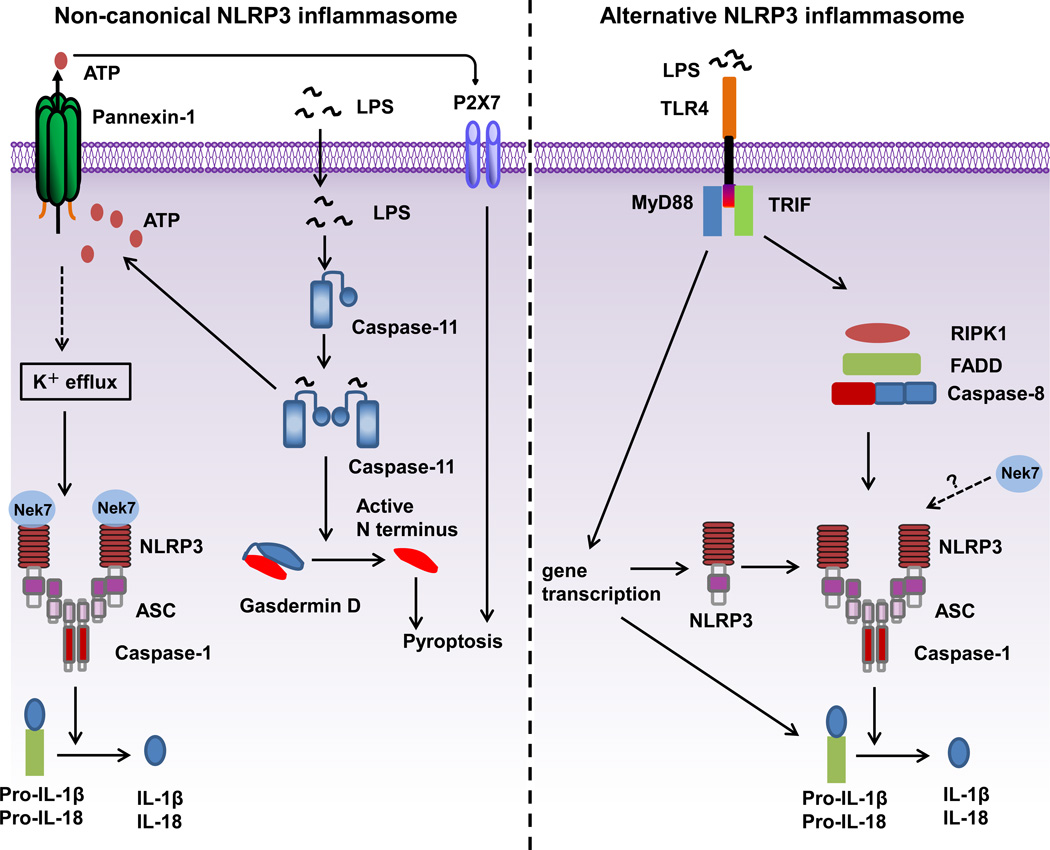
Figure [3]

Figure [4]
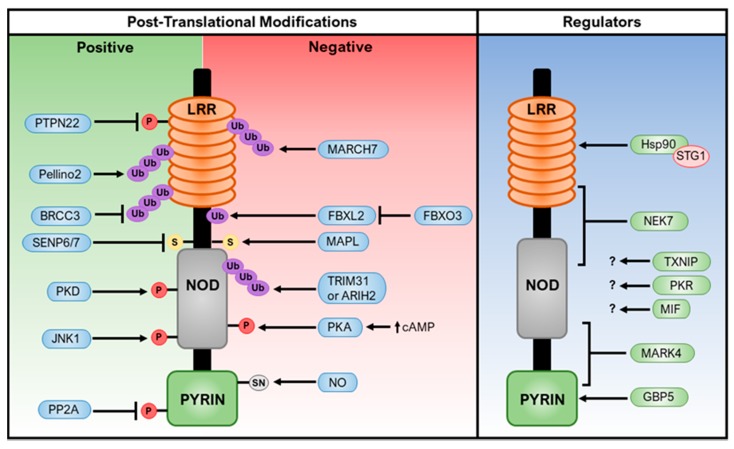
Figure [5]
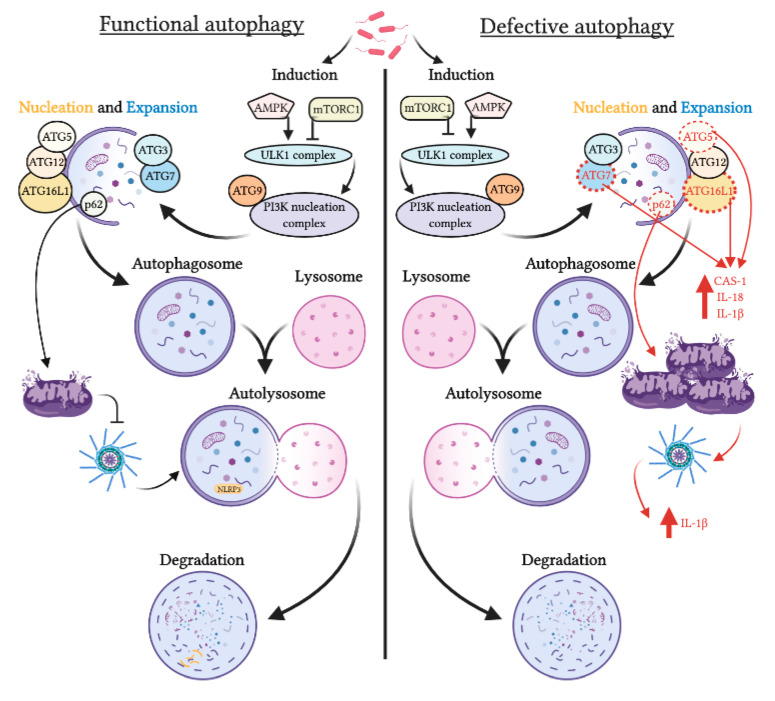
Figure [6]

Figure [7]

Figure [8]
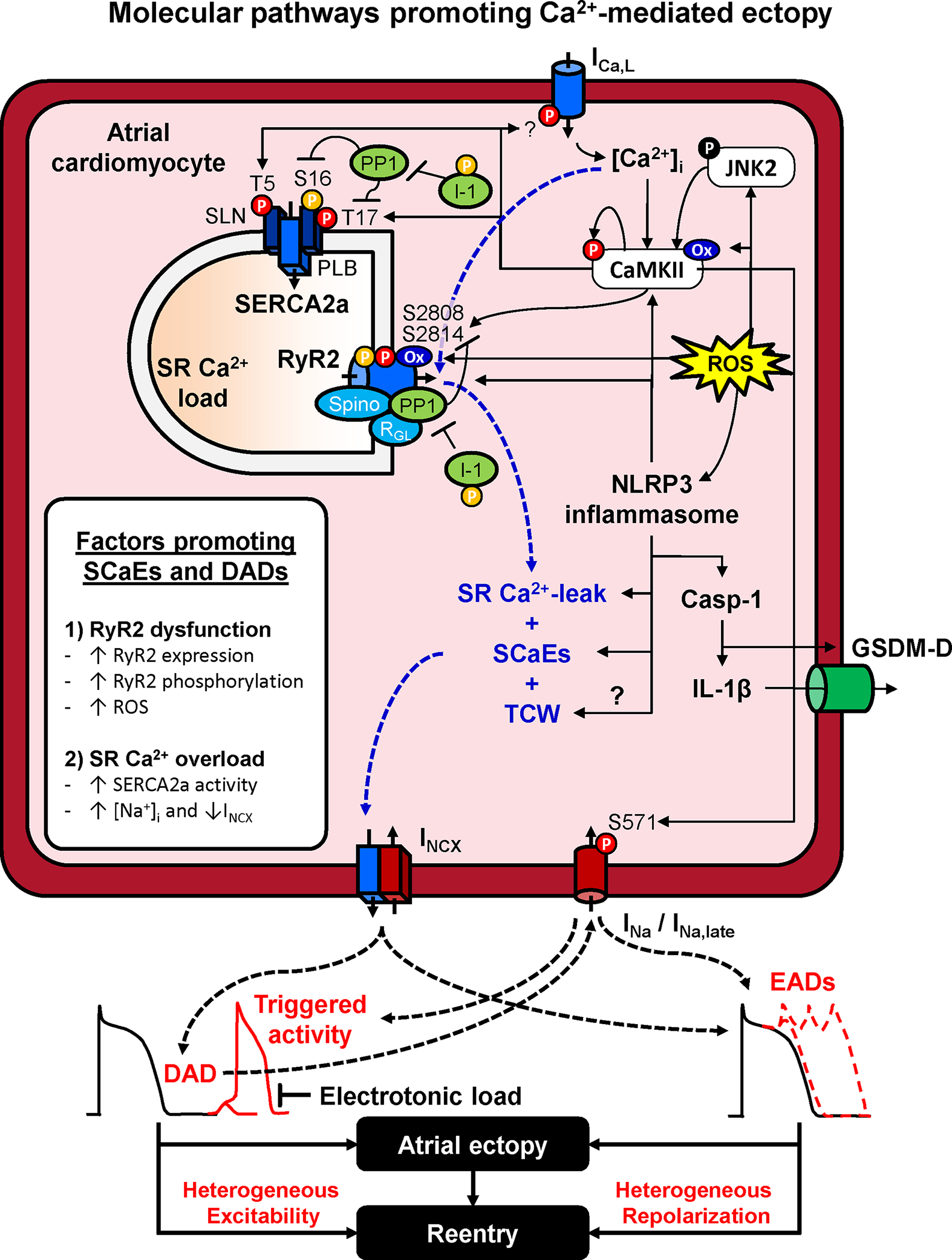
Figure [9]
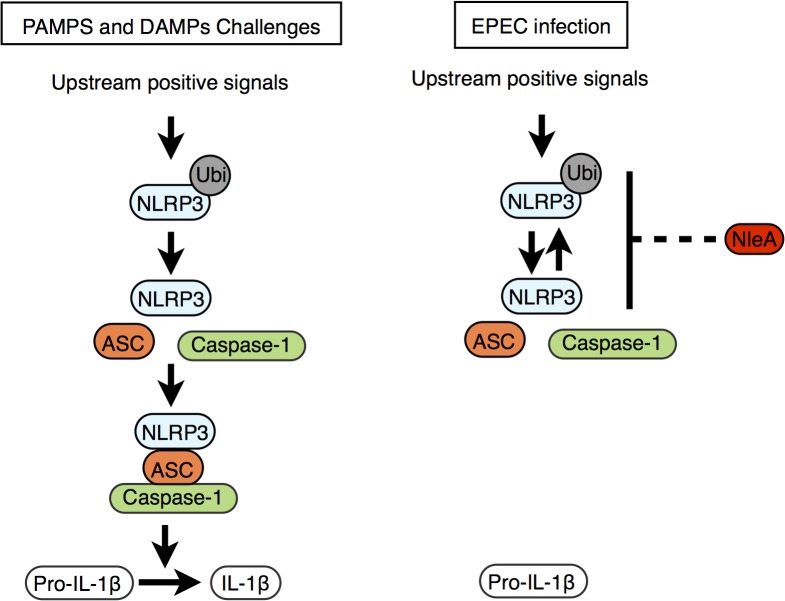
Figure [10]

Note: If you are interested in the full version of this target analysis report, or if you'd like to learn how our AI-powered BDE-Chem can design therapeutic molecules to interact with the NLRP3 target at a cost 90% lower than traditional approaches, please feel free to contact us at BD@silexon.ai.
More Common Targets
ABCB1 | ABCG2 | ACE2 | AHR | AKT1 | ALK | AR | ATM | BAX | BCL2 | BCL2L1 | BECN1 | BRAF | BRCA1 | CAMP | CASP3 | CASP9 | CCL5 | CCND1 | CD274 | CD4 | CD8A | CDH1 | CDKN1A | CDKN2A | CREB1 | CXCL8 | CXCR4 | DNMT1 | EGF | EGFR | EP300 | ERBB2 | EREG | ESR1 | EZH2 | FN1 | FOXO3 | HDAC9 | HGF | HMGB1 | HSP90AA1 | HSPA4 | HSPA5 | IDO1 | IFNA1 | IGF1 | IGF1R | IL17A | IL6 | INS | JUN | KRAS | MAPK1 | MAPK14 | MAPK3 | MAPK8 | MAPT | MCL1 | MDM2 | MET | MMP9 | MTOR | MYC | NFE2L2 | NLRP3 | NOTCH1 | PARP1 | PCNA | PDCD1 | PLK1 | PRKAA1 | PRKAA2 | PTEN | PTGS2 | PTK2 | RELA | SIRT1 | SLTM | SMAD4 | SOD1 | SQSTM1 | SRC | STAT1 | STAT3 | STAT5A | TAK1 | TERT | TLR4 | TNF | TP53 | TXN | VEGFA | YAP1

