ABCG2 Target Analysis Report Summary


About the Target
Based on the context information provided, ABCG2 (also known as ABCB2) is a gene that plays a significant role in drug resistance in various types of cancer cells. Several models have been proposed to explain the mechanisms underlying ABCG2-mediated resistance in different contexts.
CDK7i resistance in triple-negative breast cancer (TNBC) cells: Increased TGF-beta/activin signaling in CDK7i-resistant TNBC cells leads to increased expression of ABCG2 and EMT. Inhibiting TGF-beta/activin signaling or ABCG2 can resensitize the CDK7i-resistant cells to treatment [1].
Corneal epithelial stem cells: Substrate stiffness affects the behavior of corneal epithelial stem cells. On soft substrates, ABCG2 is highly expressed, contributing to stem cell maintenance. However, substrate stiffening suppresses ABCG2 expression and promotes cell differentiation [2].
5-FU-resistant colorectal cancer (CRC) cells: The miR-21/PDCD4/JNK/ABCG2 axis is a major molecular mechanism in 5-FU-resistant CRC cells. Understanding this axis may provide insights into combating drug resistance [3].
Multidrug resistance (MDR) in cancer cells: Overexpression of ABCG2 leads to the efflux of antitumor drugs, resulting in drug resistance. CM082, an inhibitor of ABCG2, can increase drug accumulation in tumor cells and restore drug sensitivity [4].
Gemcitabine (GEM) resistance in pancreatic cancer cells: PRMT3, when upregulated in GEM-resistant pancreatic cancer cells, stabilizes ABCG2 mRNA through methylation. This upregulation of ABCG2 contributes to gemcitabine resistance [5].
In summary, ABCG2 is involved in drug resistance in various cancer types through different molecular mechanisms. Modulating ABCG2 expression or activity, either directly or indirectly, may be a viable approach for overcoming drug resistance in these contexts. [1][2][3][4][5].
Based on the context information provided, the following key viewpoints can be extracted:
The transporters and enzymes involved in the absorption, distribution, metabolism, and excretion of rosuvastatin were incorporated into a model of various organs and tissues [6].
The model illustrated the main sites of action for these transporters and enzymes, including the liver for Pgp and the kidney for BCRP [6].
UCP2, a protein involved in regulating cellular metabolism, is downregulated by hypoxia [7].
Hypoxia-induced downregulation of UCP2 is linked to chemoresistance in NSCLC cells through the upregulation of ABCG2 [7].
UCP2 knockdown increases glucose uptake and lactate production, while UCP2 overexpression decreases glucose consumption and lactate production [7].
Hypoxic cells show decreased amounts of components involved in oxidative phosphorylation complexes [7].
UCP2 downregulation promotes the efflux of chemotherapeutic drugs through the upregulation of ABCG2 [7].
These viewpoints provide a comprehensive summary of the relationship between UCP2, ABCG2, and cellular metabolism under hypoxic conditions.
Figure [1]

Figure [2]

Figure [3]

Figure [4]

Figure [5]

Figure [6]
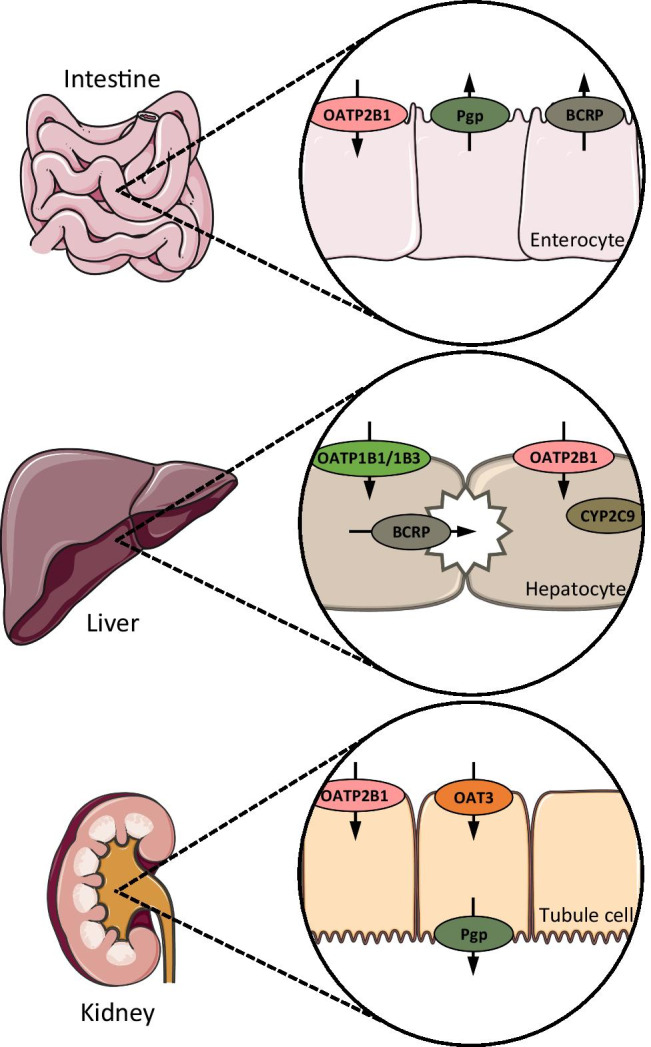
Figure [7]

Note: If you are interested in the full version of this target analysis report, or if you'd like to learn how our AI-powered BDE-Chem can design therapeutic molecules to interact with the ABCG2 target at a cost 90% lower than traditional approaches, please feel free to contact us at BD@silexon.ai.
More Common Targets
ABCB1 | ABCG2 | ACE2 | AHR | AKT1 | ALK | AR | ATM | BAX | BCL2 | BCL2L1 | BECN1 | BRAF | BRCA1 | CAMP | CASP3 | CASP9 | CCL5 | CCND1 | CD274 | CD4 | CD8A | CDH1 | CDKN1A | CDKN2A | CREB1 | CXCL8 | CXCR4 | DNMT1 | EGF | EGFR | EP300 | ERBB2 | EREG | ESR1 | EZH2 | FN1 | FOXO3 | HDAC9 | HGF | HMGB1 | HSP90AA1 | HSPA4 | HSPA5 | IDO1 | IFNA1 | IGF1 | IGF1R | IL17A | IL6 | INS | JUN | KRAS | MAPK1 | MAPK14 | MAPK3 | MAPK8 | MAPT | MCL1 | MDM2 | MET | MMP9 | MTOR | MYC | NFE2L2 | NLRP3 | NOTCH1 | PARP1 | PCNA | PDCD1 | PLK1 | PRKAA1 | PRKAA2 | PTEN | PTGS2 | PTK2 | RELA | SIRT1 | SLTM | SMAD4 | SOD1 | SQSTM1 | SRC | STAT1 | STAT3 | STAT5A | TAK1 | TERT | TLR4 | TNF | TP53 | TXN | VEGFA | YAP1

