HSPA5 Target Analysis Report Summary


About the Target
HSPA5, also known as GRP78 or BiP, is a crucial protein involved in various cellular processes. It is primarily found in the endoplasmic reticulum (ER) where it plays a role in the activation of the unfolded protein response (UPR), ER-associated degradation (ERAD), and regulation of the mitochondria-associated ER membrane (MAM) [1].
GRP78 can be regulated at different levels, including transcriptional regulation mediated by various transcription factors binding to specific motifs in its gene promoter [1]. Additionally, alternative processing of GRP78's pre-mRNA can occur under stressful conditions, resulting in the production of a truncated protein that is retained in the cytosol [1]. Post-transcriptional regulation of GRP78 also occurs through the action of factors on its IRES motif or the action of different miRNAs [1].
In response to ER conditions, GRP78's activity can be modulated by the protein HYPE, which AMPylates or deAMPylates GRP78 depending on the unfolded protein load in the ER [2]. AMPylated GRP78 is non-functional and pooled into a reservoir, while deAMPylated GRP78 can be activated and function as a chaperone under stress conditions [2]. This switch between AMPylation and deAMPylation states is mediated by specific residues and the interaction with Arg374 and a water molecule [2].
Furthermore, GRP78 interacts with misfolded proteins, as shown through its interaction with misfolded antitrypsin with the UPR sensor IRE1alpha [3]. This interaction leads to the release of BiP from UPR sensors and subsequent activation of UPR signaling pathways [3]. Misfolded proteins can also induce conformational changes in UPR sensors, such as IRE1alpha and PERK, which activate their kinase and RNase activities [3]. ATF6alpha complexes may undergo disassembly in response to misfolded proteins, migrating to the Golgi for proteolytic processing [3].
In a different context, GRP78 is implicated in carfilzomib-induced cardiotoxicity, with metformin shown to have a prophylactic effect [4]. In aged myocardium, the induction of autophagy is recognized as a cardioprotective strategy [4]. The combined effect of carfilzomib and metformin resulted in the synergistic induction of autophagy, leading to improved left ventricular function and establishing metformin as a potential prophylactic therapy in aged individuals [4].
Finally, GRP78's involvement in the canonical Wnt signaling pathway has been investigated [5]. It was found that GRP78 interacts with DCD at the cell surface, regulating cell migration by activating the canonical Wnt signaling pathway [5]. This interaction leads to the activation of beta-catenin transcriptional activity and increased expression of Wnt target genes [5]. The proposed model suggests that GRP78 and DCD interaction influences cell migration and the Wnt signaling pathway [5].
Overall, GRP78 (HSPA5/BiP) is a multifunctional protein involved in UPR, ERAD, MAM regulation, autophagy induction, and Wnt signaling pathway activation. Its regulation occurs at different levels, including transcriptional and post-transcriptional regulation. Its interaction with HYPE, misfolded proteins, and DCD contributes to its diverse functions within the cell.
Based on the given context information, here is a comprehensive summary of key viewpoints regarding HSPA5 (also known as GRP78):
HSPA5 is involved in protein translocation into the endoplasmic reticulum (ER) and the mitochondrial matrix [6].
HSPA5 acts as a driving force for polypeptide insertion into the ER lumen, facilitating the translocation process [6].
In the ER, HSPA5 binds to nascent polypeptide chains, increasing their entropy as they move away from the membrane constraints [6].
Allosteric inhibition of HSPA5's activity by YUM70 leads to increased endoplasmic reticulum (ER) stress and prolonged ER stress-mediated apoptosis in pancreatic cancer cells [7].
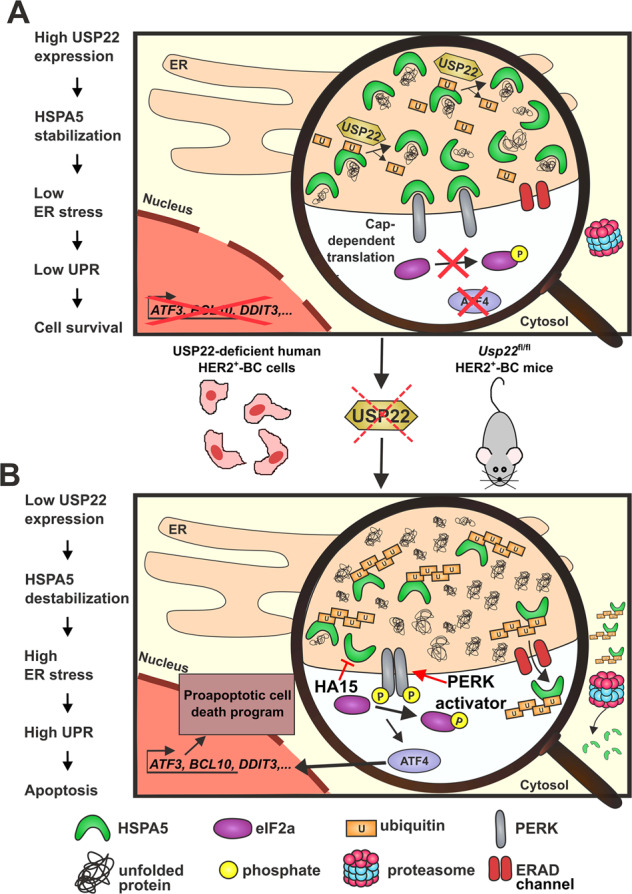
HSPA5 is stabilized by USP22, which enhances the protein folding capacity in the ER and maintains low levels of unfolded protein response (UPR) signaling in cancer cells [8].
Impaired expression of USP22 leads to decreased HSPA5 protein stability, resulting in the accumulation of unfolded proteins in the ER and stimulation of UPR signaling [8].
Activation of the PERK/ATF4/ATF3 axis in response to UPR induction leads to programmed cell death in a p53-independent manner [8].
Inhibitors targeting HSPA5, such as HA15, and the PERK activator CCT020312 have been developed and may contribute to the development of new UPR-based anti-cancer therapeutic strategies [8].
Impaired expression of USP22 sensitizes HER2+-BC (HER2-positive breast cancer) to programmed cell death along the HSPA5/PERK/ATF4/ATF3-axis of the UPR [8].
References:
[6] Protein translocation and its driving force - Information from source [6]
[7] YUM70-mediated cell death through inhibition of GRP78 - Information from source [7]
[8] Role of HSPA5 and USP22 in UPR signaling and cell death - Information from source [8]
Figure [1]
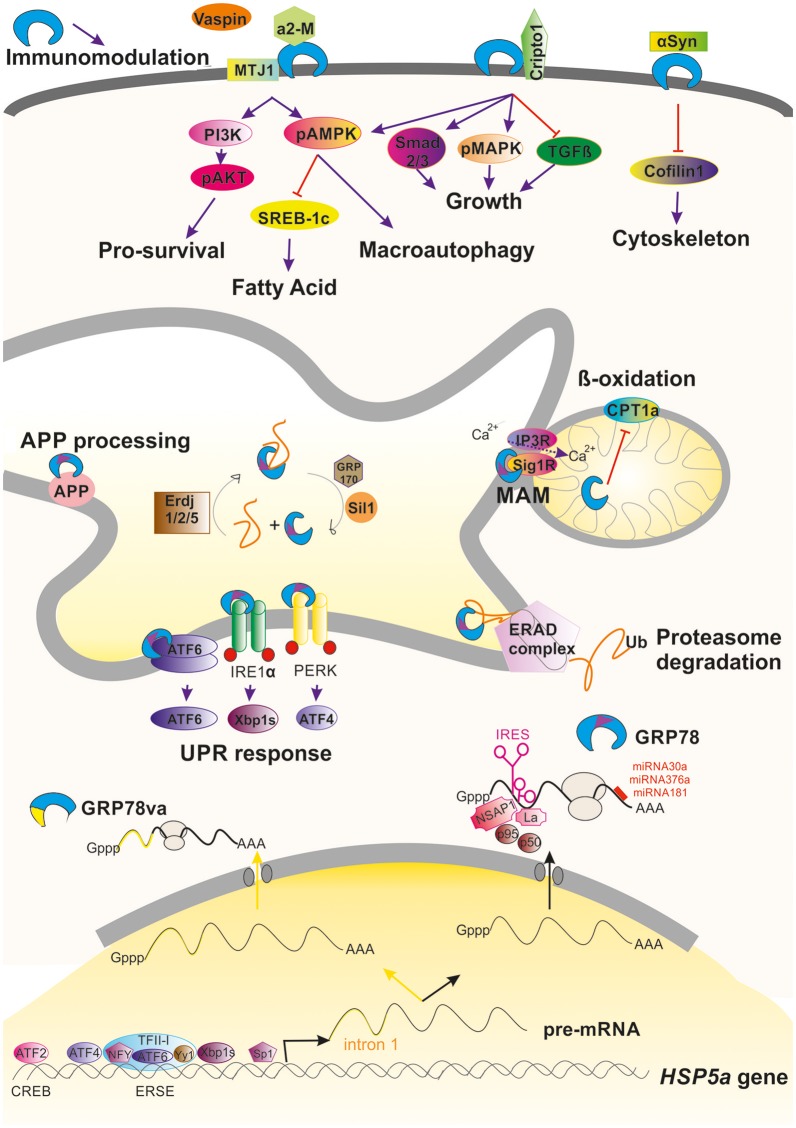
Figure [2]
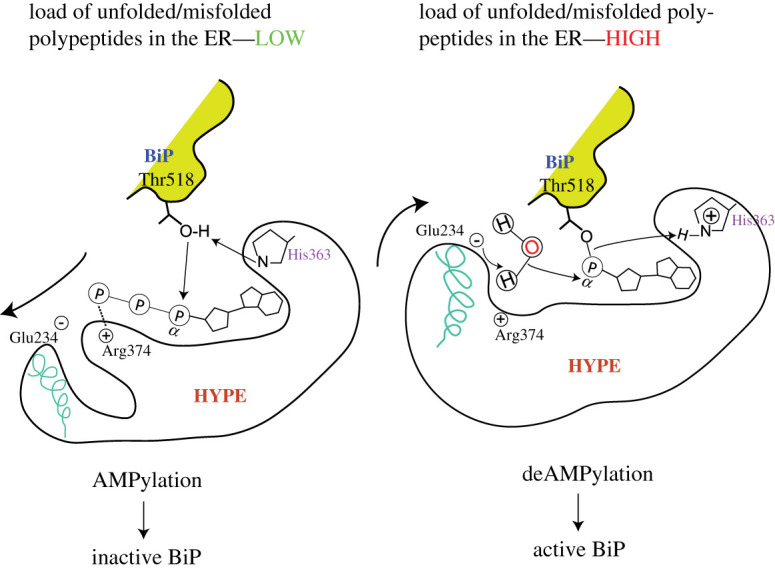
Figure [3]
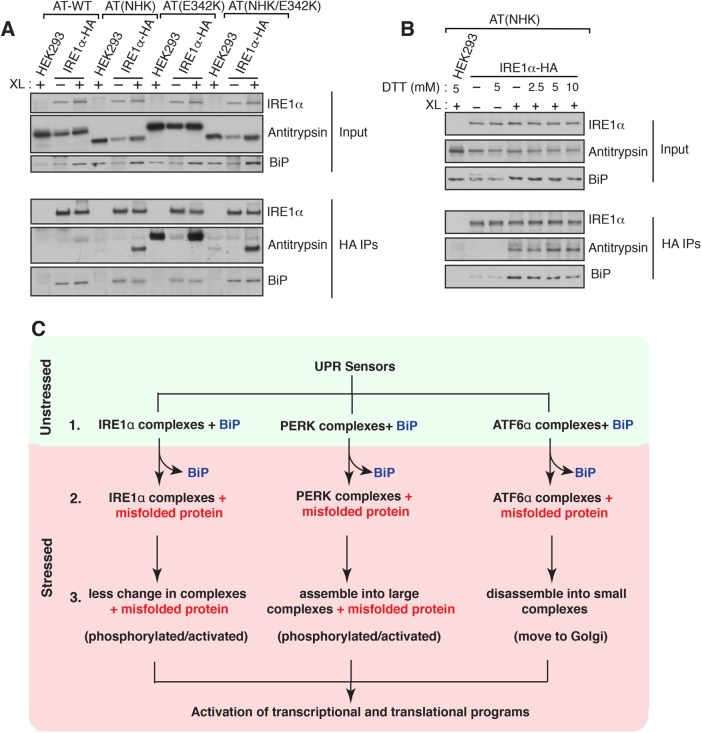
Figure [4]

Figure [5]

Figure [6]
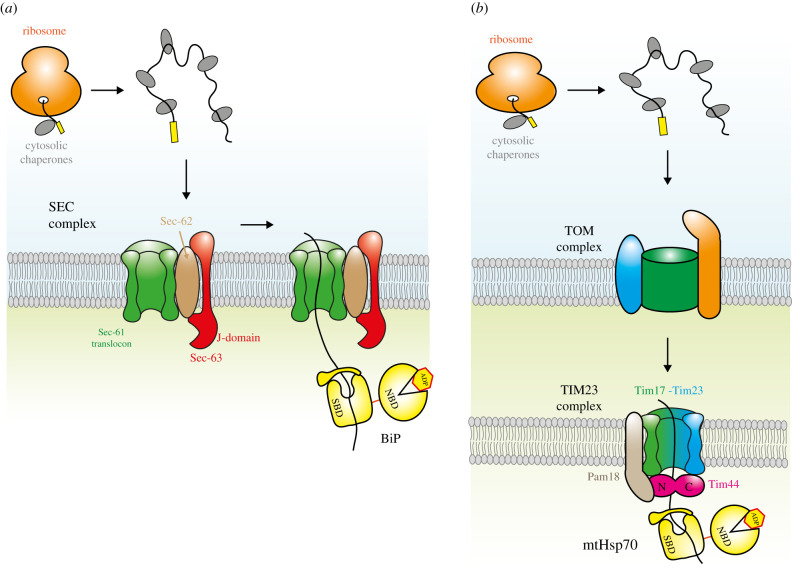
Figure [7]
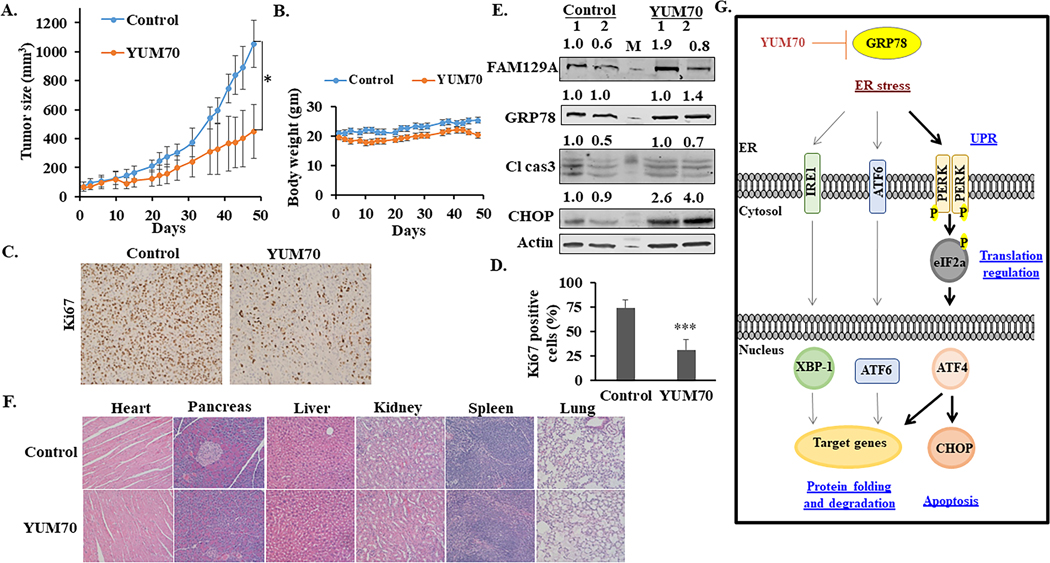
Figure [8]
Note: If you are interested in the full version of this target analysis report, or if you'd like to learn how our AI-powered BDE-Chem can design therapeutic molecules to interact with the HSPA5 target at a cost 90% lower than traditional approaches, please feel free to contact us at BD@silexon.ai.
More Common Targets
ABCB1 | ABCG2 | ACE2 | AHR | AKT1 | ALK | AR | ATM | BAX | BCL2 | BCL2L1 | BECN1 | BRAF | BRCA1 | CAMP | CASP3 | CASP9 | CCL5 | CCND1 | CD274 | CD4 | CD8A | CDH1 | CDKN1A | CDKN2A | CREB1 | CXCL8 | CXCR4 | DNMT1 | EGF | EGFR | EP300 | ERBB2 | EREG | ESR1 | EZH2 | FN1 | FOXO3 | HDAC9 | HGF | HMGB1 | HSP90AA1 | HSPA4 | HSPA5 | IDO1 | IFNA1 | IGF1 | IGF1R | IL17A | IL6 | INS | JUN | KRAS | MAPK1 | MAPK14 | MAPK3 | MAPK8 | MAPT | MCL1 | MDM2 | MET | MMP9 | MTOR | MYC | NFE2L2 | NLRP3 | NOTCH1 | PARP1 | PCNA | PDCD1 | PLK1 | PRKAA1 | PRKAA2 | PTEN | PTGS2 | PTK2 | RELA | SIRT1 | SLTM | SMAD4 | SOD1 | SQSTM1 | SRC | STAT1 | STAT3 | STAT5A | TAK1 | TERT | TLR4 | TNF | TP53 | TXN | VEGFA | YAP1

