SOD1 Target Analysis Report Summary


About the Target
Based on the provided context information, the key viewpoints regarding SOD1 are as follows:
SOD1 mutations, such as mSOD1, can trigger endoplasmic reticulum stress and unfolded protein response (UPR), leading to the accumulation of misfolded mutant proteins and activation of autophagy pathways [1A].
Oxidative stress and mitochondrial dysfunction, caused by mutations in SOD1, can activate autophagy and trigger mitophagy, which helps in clearing damaged mitochondria [1B].
Mutations in RNA-binding proteins, including SOD1, can lead to defects in RNA metabolism and alter the dynamics of stress granules, potentially affecting autophagy [1C].
DNA damage, either as a result of pathogenic mutations or oxidative stress, is a common feature in SOD1 mutations and can activate autophagy through the AMPK pathway [1D].
The metalation and activation of SOD1 is facilitated by the interaction with copper chaperone for SOD1 (CCS) protein, leading to the formation of fully metalated holo-SOD1 monomers that can homodimerize [2].
SOD1 regulation can be influenced by post-translational modifications (PTMs), including those driven by factors such as DNA damage, amino acid starvation, and metabolic fitness [3].
Extracellular vesicles (EVs) play a role in ALS pathology by spreading mutant proteins implicated in ALS, including mutant SOD1, TDP-43, FUS, and DPRs from expanded C9orf72 [4].
SOD1 oxidation is modulated by the levels of Cd2+, Zn2+, and metallothioneins (MTs), with excess Cd2+ causing redox imbalance and oxidation of SOD1, while supplementation of excess Zn2+ and induction of MTs can prevent SOD1 oxidation [5].
References:
[1] Cross-talk between autophagy and other pathogenic mechanisms in amyotrophic lateral sclerosis (ALS)
[2] Transitional free-energy landscapes of SOD1 metalloforms and CCS-dependent SOD1 maturation
[3] PTM-driven mechanisms of SOD1 regulation
[4] EVs in ALS pathology
[5] Cd2+-induced SOD1 oxidation is modulated by Zn2+ and MT levels
Based on the provided context information, the following key viewpoints can be extracted regarding SOD1:
In prion-like diseases, like ALS, the misfolding of SOD1 leads to a reduction in protein function, which may contribute to the pathology [6].
Normally folded prion protein (PrPC) is reduced in abundance in prion disease paradigms, and this reduction may be linked to the neuroprotection functions associated with PrPC [6].
Misfolding of proteins like PrPC and SOD1 can also impact the functions of the proteins they interact with, which may be neuroprotective or essential for neuron survival [6].
It is unclear if the abundance of normally folded SOD1 decreases with disease progression, but misfolded SOD1 conformers with reduced function could explain the loss-of-function component in ALS pathology [6].
SOD1 plays a role in the detoxification of reactive oxygen species (ROS), and its upregulation, along with other proteins like Prdx1, contributes to the lifespan of red blood cells [7].
In glioblastoma-derived cells, the interaction between neurotransmitter substance P (SP) and its receptor NK1R can lead to an increase in intracellular ROS levels by suppressing the enzymatic activities of catalase and SOD. However, blocking NK1R using a drug called aprepitant can restore the oxidative balance and reduce the survival and proliferative capacity of these cells [8].
ALS-related SOD1 G93A mutation impairs the binding of VDAC1 and HK1, leading to mitochondrial dysfunction. However, a peptide called NHK1 can inhibit the binding of SOD1 G93A to VDAC1 and improve mitochondrial function and cell viability [9].
In patients with oral cancer, lower SOD enzyme activity has been reported, suggesting a potential role of SOD in the progression of oral cancer [8].
References:
[6] Context Information, [7] Context Information, [8] Context Information, [9] Context Information.
Figure [1]

Figure [2]

Figure [3]

Figure [4]
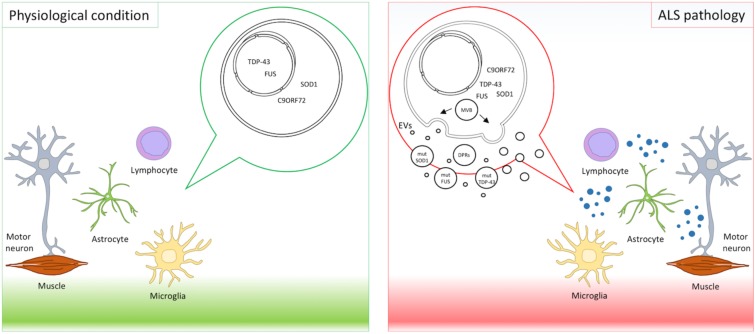
Figure [5]

Figure [6]

Figure [7]

Figure [8]
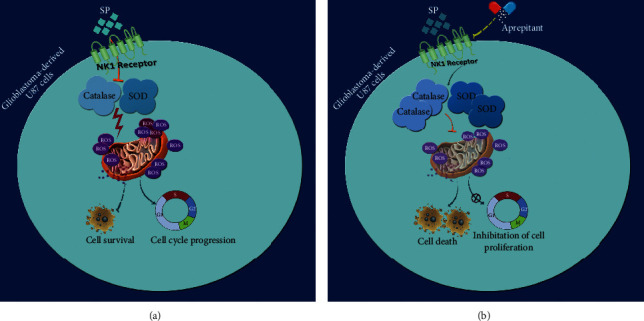
Figure [9]

Note: If you are interested in the full version of this target analysis report, or if you'd like to learn how our AI-powered BDE-Chem can design therapeutic molecules to interact with the SOD1 target at a cost 90% lower than traditional approaches, please feel free to contact us at BD@silexon.ai.
More Common Targets
ABCB1 | ABCG2 | ACE2 | AHR | AKT1 | ALK | AR | ATM | BAX | BCL2 | BCL2L1 | BECN1 | BRAF | BRCA1 | CAMP | CASP3 | CASP9 | CCL5 | CCND1 | CD274 | CD4 | CD8A | CDH1 | CDKN1A | CDKN2A | CREB1 | CXCL8 | CXCR4 | DNMT1 | EGF | EGFR | EP300 | ERBB2 | EREG | ESR1 | EZH2 | FN1 | FOXO3 | HDAC9 | HGF | HMGB1 | HSP90AA1 | HSPA4 | HSPA5 | IDO1 | IFNA1 | IGF1 | IGF1R | IL17A | IL6 | INS | JUN | KRAS | MAPK1 | MAPK14 | MAPK3 | MAPK8 | MAPT | MCL1 | MDM2 | MET | MMP9 | MTOR | MYC | NFE2L2 | NLRP3 | NOTCH1 | PARP1 | PCNA | PDCD1 | PLK1 | PRKAA1 | PRKAA2 | PTEN | PTGS2 | PTK2 | RELA | SIRT1 | SLTM | SMAD4 | SOD1 | SQSTM1 | SRC | STAT1 | STAT3 | STAT5A | TAK1 | TERT | TLR4 | TNF | TP53 | TXN | VEGFA | YAP1

