IGF1R Target Analysis Report Summary


About the Target
Based on the provided context information, here is a comprehensive summary of key viewpoints related to IGF1R:
IGF-1R signaling plays a crucial role in promoting cell proliferation, growth, and survival in differentiated normal cells. It also promotes mesenchymal stem cell proliferation and multiple lineage differentiation through interaction with other signaling pathways [1].
In embryonic and germline stem cells, autocrine/paracrine IGF-1R signaling is important for maintaining stem cell survival and pluripotency status. Niche cells in the microenvironment contribute to this regulation [1].
Niche hypoxia can activate IGF-1R signaling in germline stem cells, promoting their self-renewal and migration [1].
In liver cancer, epigenetic alterations and niche inflammation lead to upregulation of IGF-1R signaling. This pathway also contributes to stemness in liver cancer stem cells and niche inflammation [1].
MiR-12528 can negatively regulate the development and progression of non-small cell lung cancer (NSCLC) by blocking IGF-1R translation, which reduces the interaction between IGF-1R and its ligand. This, in turn, affects cell cycle regulation and programmed cell death [2].
High expression of the m6a reader YTHDC2 in nasopharyngeal carcinoma cells can lead to increased translation efficiency of IGF1R mRNA, resulting in resistance to irradiation [3].
Mutations in the tumor suppressor gene p53, which is frequently mutated in human cancer, can lead to increased concentrations of IGF1R mRNA and cell-surface receptors. This promotes uncontrolled proliferation and abrogation of apoptosis in tumor cells [4].
IGFBP-5 and the metalloproteinase Papp-aa act as part of a molecular switch that controls IGF signaling in target cells. Under normal conditions, IGFBP-5 inhibits IGF signaling by binding to IGFs and preventing their binding to IGF-1R. However, under low calcium conditions, increased Papp-aa activity leads to IGFBP-5a cleavage, releasing IGFs and activating IGF-1 receptor-mediated signaling [5].
These viewpoints highlight the diverse roles of IGF1R signaling in various cell types and cancers, as well as the influence of the microenvironment and regulatory factors.
Based on the provided information, several key viewpoints regarding IGF1R can be summarized as follows:
IGF1R plays a role in the crosstalk between thyroid-stimulating hormone receptor (TSHR) and hyaluronan secretion in Graves' orbital fibroblasts, with different phases of hyaluronan secretion being observed in response to M22 stimulation [6].
In chronic lymphocytic leukemia (CLL), EZH2 may regulate the PI3K pathway through IGF1R, potentially affecting the activation of the pathway in different types of CLL [7].
The canonical signaling pathway of IGF1R involves the binding of IGF-I and IGF-II to the extracellular alpha subunit of IGF1R, leading to the activation of downstream signaling proteins such as IRS and Akt. This pathway regulates apoptosis, cell metabolism, protein synthesis, and cell proliferation, among other cellular processes [8].
Besides the canonical pathway, the activated IGF1R and insulin receptor (IR) can interact with several other factors, including DDRs, FAK, Src, MET, RON, JAK, and STAT. These interactions contribute to various cellular processes such as autophagy, epithelial-mesenchymal transition (EMT), stemness, and anoikis [8].
The GH-IGF-I axis is involved in the regulation of IGF-I production and bioavailability. Mutations in components of this axis, such as STAT5B, IGF1, IGFALS, PAPPA2, and IGF1R, can lead to growth hormone insensitivity (GHI), IGF deficiency, altered IGF bioavailability, or IGF resistance, which can have associated co-morbidities [9].
The IGF-1R pathway can be activated by DNA damaging stress and inhibited by AKT. AKT plays a role in inhibiting p53-dependent apoptosis, while also promoting p53 protein synthesis through mTORC1 activity. Inhibitors of IGF-1R and AKT can increase p53-dependent apoptosis and reduce p53-mediated senescence [10].
References:
[6] Context unknown
[7] Context unknown
[8] Context unknown
[9] Context unknown
[10] Context unknown
Figure [1]

Figure [2]

Figure [3]

Figure [4]

Figure [5]

Figure [6]
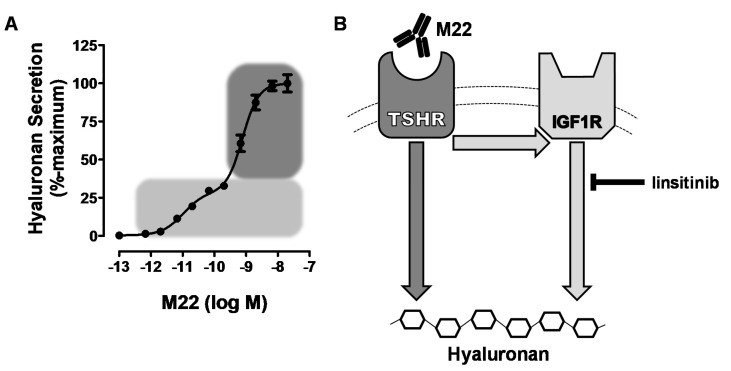
Figure [7]

Figure [8]
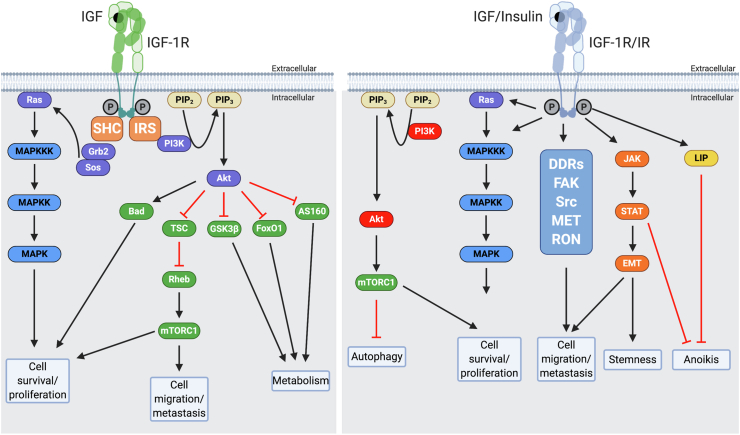
Figure [9]

Figure [10]

Note: If you are interested in the full version of this target analysis report, or if you'd like to learn how our AI-powered BDE-Chem can design therapeutic molecules to interact with the IGF1R target at a cost 90% lower than traditional approaches, please feel free to contact us at BD@silexon.ai.
More Common Targets
ABCB1 | ABCG2 | ACE2 | AHR | AKT1 | ALK | AR | ATM | BAX | BCL2 | BCL2L1 | BECN1 | BRAF | BRCA1 | CAMP | CASP3 | CASP9 | CCL5 | CCND1 | CD274 | CD4 | CD8A | CDH1 | CDKN1A | CDKN2A | CREB1 | CXCL8 | CXCR4 | DNMT1 | EGF | EGFR | EP300 | ERBB2 | EREG | ESR1 | EZH2 | FN1 | FOXO3 | HDAC9 | HGF | HMGB1 | HSP90AA1 | HSPA4 | HSPA5 | IDO1 | IFNA1 | IGF1 | IGF1R | IL17A | IL6 | INS | JUN | KRAS | MAPK1 | MAPK14 | MAPK3 | MAPK8 | MAPT | MCL1 | MDM2 | MET | MMP9 | MTOR | MYC | NFE2L2 | NLRP3 | NOTCH1 | PARP1 | PCNA | PDCD1 | PLK1 | PRKAA1 | PRKAA2 | PTEN | PTGS2 | PTK2 | RELA | SIRT1 | SLTM | SMAD4 | SOD1 | SQSTM1 | SRC | STAT1 | STAT3 | STAT5A | TAK1 | TERT | TLR4 | TNF | TP53 | TXN | VEGFA | YAP1

