EGFR Target Analysis Report Summary


About the Target
Based on the provided context information, here is a comprehensive summary of key viewpoints related to EGFR:
Location and Regulation of HER Signaling: HER signaling occurs in various cellular compartments, including the plasma membrane, cytoplasm, and nucleus. Understanding the regulation of HER activity in different levels provides versatile approaches to target HER signaling in Head and Neck Cancer (HNC) [1].
Targeting HER Signaling in HNC: HER signaling in HNC can be targeted horizontally by blocking dimerization and transactivation between different HER members and/or their association with other RTKs, or vertically by targeting different signaling nodes, endocytosis/trafficking pathways, and nuclear translocation [1].
Poly-Therapeutic Strategies: Combined targeting of YAP (a component of the Hippo pathway) and EGFR signaling may enhance the efficacy of Hepatocellular Carcinoma (HCC) treatment [2].
Deep Learning for Signaling Pathways: Deep Learning Models (DLMs) can be used to infer the activation states of signaling pathways, including the EGFR signaling cascade, by learning from gene expression data [3].
Functions of EGFR in Cancer and COVID-19: EGFR plays a role in promoting proliferation, inflammation, angiogenesis, fibrosis, epithelial-mesenchymal transition, and metastasis in cancer. In COVID-19, EGFR contributes to inflammation and fibrosis [4].
Targeting EGFR in HNC: Combining targeting of EGFR central signaling and cancer metabolism may be a rational treatment strategy for HNC, offering a multilateral approach against the disease [5].
These viewpoints illustrate the diverse roles of EGFR in cellular signaling, its potential as a therapeutic target, and its involvement in cancer progression and response to treatment.
Based on the provided context information, here is a comprehensive summary of the key viewpoints related to EGFR:
Antibody-dependent cell-mediated cytotoxicity (ADCC) mediated by cetuximab, an EGFR-targeting antibody, plays a role in the cytotoxic effect against cancer cells. This effect involves the activation of NK cells and the interaction between cetuximab-activated NK cells and cancer cells through NKG2D-MICA binding. This interaction leads to dendritic cell maturation and enhances cytolytic T lymphocyte activity towards cancer cells [6B].
EGFR-JAK2-STAT3 signaling pathway activation can result in increased levels of immune suppressive cytokines IL-6, IL-10, and TGF-beta. These cytokines, along with IL-10 and TGF-beta, can activate regulatory T cells (Treg cells) which suppress the cytotoxic effects of NK cells and CD8+ T cells [6A].
Inhibition of CD8+ T cells by Treg cells, which are activated through the increased levels of IL-6, IL-10, and TGF-beta, may contribute to the protection of cancer cells against the cytotoxic effects of cetuximab [6A].
The EGFR T790M mutation can activate NADPH oxidase, leading to the production of reactive oxygen species (ROS) and mild oxidation of Met790. In normal conditions, Met reductase A (MsrA) can reverse this oxidation and protect the EGFR from degradation, promoting cell survival. However, in conditions of excessive ROS production, such as sanguinarine-mediated ROS overproduction, the redox imbalance caused by NOX3 activation leads to MsrA inactivation and further oxidation of Met790. This ultimately results in EGFR degradation and cell apoptosis [7].
The mechanisms of acquired resistance to third-generation EGFR-TKIs can be classified as EGFR-dependent and EGFR-independent. EGFR-dependent resistance involves EGFR re-mutation, T790M deletion, and EGFR amplification. EGFR-independent resistance mechanisms include MET and HER2 amplification, RAS and BRAF mutation, histological transformation, EMT, oncogene fusion, activation of the PI3K/Akt/mTOR pathway, PTEN deletion, FGFR signaling pathway abnormalities, and induction of angiogenesis [8].
The activation of the EGFR signaling pathway, fueled by the expression of EGFR ligands and EGFR activation, contributes to basal HNSCC cell proliferation and survival. Blockade of EGFR with monoclonal antibodies or TKIs can down-regulate ligand gene expression and reduce the activity of the EGFR signaling pathway, thereby decreasing cell viability [9].
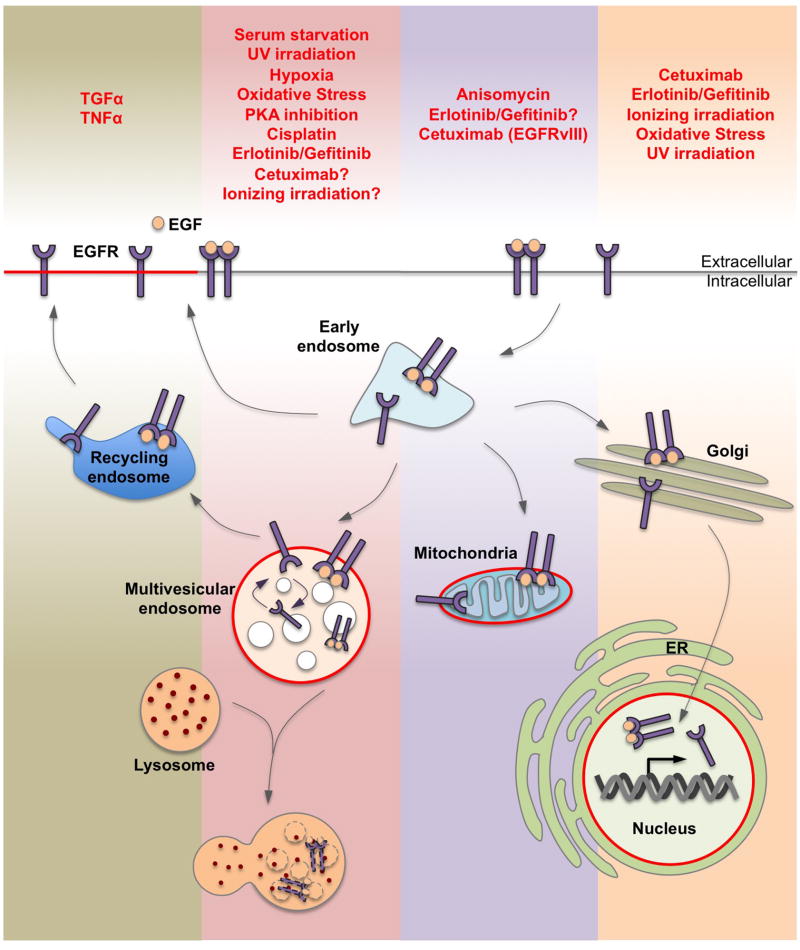
Ligand and various cellular stresses can induce EGFR internalization and trafficking. Ligand-stimulated EGFR can be recycled back to the plasma membrane or sorted to multivesicular endosomes, mitochondria, ER, and nuclei. Different cellular stresses may lead to EGFR trafficking to specific subcellular destinations. Ligand-stimulated EGFR trafficking to multivesicular endosomes targets the receptor for lysosomal degradation, while certain stresses cause perinuclear arrest of EGFR at non-degradative multivesicular endosomes. EGFR has been shown to translocate to mitochondria and potentially regulate mitochondrial pathways related to cell survival and resistance to EGFR targeting therapies [10].
Please note that these summaries are based on the provided context and demonstrate different information related to EGFR.
Figure [1]
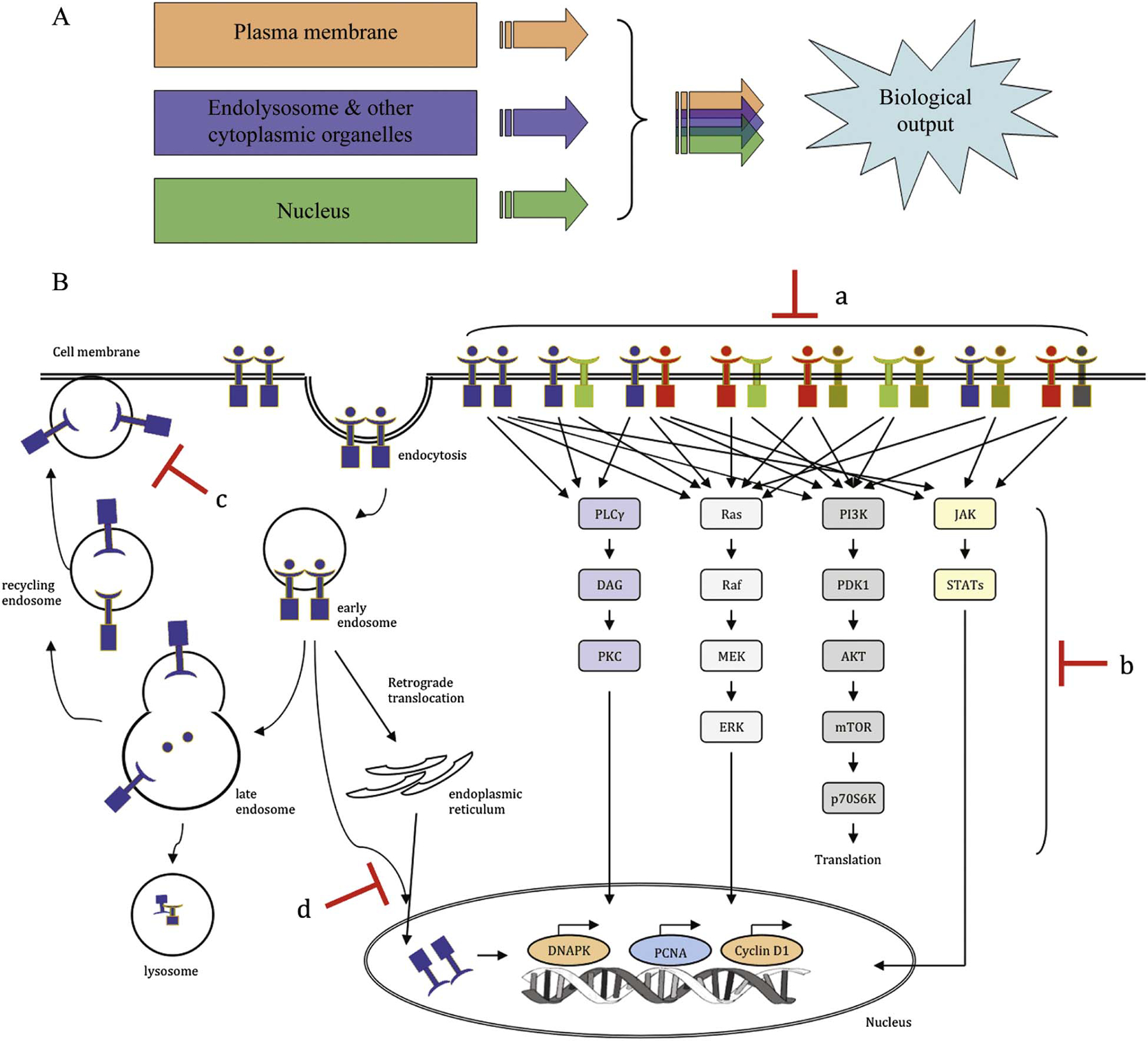
Figure [2]

Figure [3]

Figure [4]

Figure [5]

Figure [6]

Figure [7]
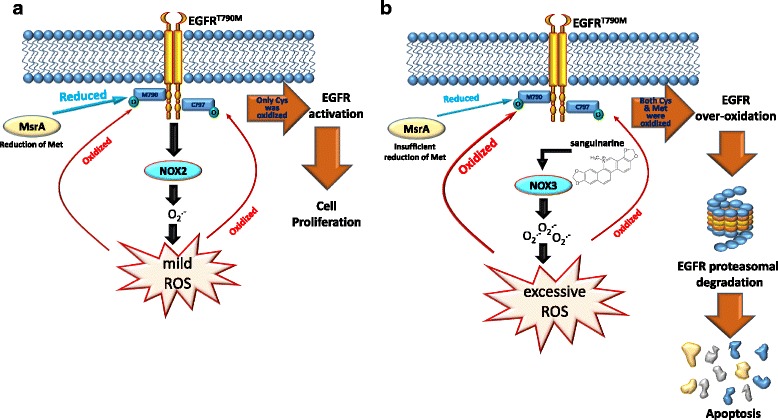
Figure [8]

Figure [9]
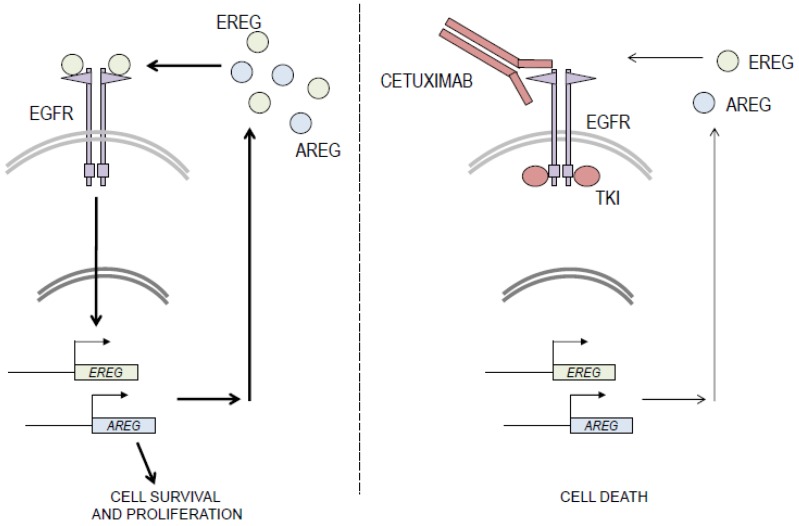
Figure [10]
Note: If you are interested in the full version of this target analysis report, or if you'd like to learn how our AI-powered BDE-Chem can design therapeutic molecules to interact with the EGFR target at a cost 90% lower than traditional approaches, please feel free to contact us at BD@silexon.ai.
More Common Targets
ABCB1 | ABCG2 | ACE2 | AHR | AKT1 | ALK | AR | ATM | BAX | BCL2 | BCL2L1 | BECN1 | BRAF | BRCA1 | CAMP | CASP3 | CASP9 | CCL5 | CCND1 | CD274 | CD4 | CD8A | CDH1 | CDKN1A | CDKN2A | CREB1 | CXCL8 | CXCR4 | DNMT1 | EGF | EGFR | EP300 | ERBB2 | EREG | ESR1 | EZH2 | FN1 | FOXO3 | HDAC9 | HGF | HMGB1 | HSP90AA1 | HSPA4 | HSPA5 | IDO1 | IFNA1 | IGF1 | IGF1R | IL17A | IL6 | INS | JUN | KRAS | MAPK1 | MAPK14 | MAPK3 | MAPK8 | MAPT | MCL1 | MDM2 | MET | MMP9 | MTOR | MYC | NFE2L2 | NLRP3 | NOTCH1 | PARP1 | PCNA | PDCD1 | PLK1 | PRKAA1 | PRKAA2 | PTEN | PTGS2 | PTK2 | RELA | SIRT1 | SLTM | SMAD4 | SOD1 | SQSTM1 | SRC | STAT1 | STAT3 | STAT5A | TAK1 | TERT | TLR4 | TNF | TP53 | TXN | VEGFA | YAP1

