MAPK3 Target Analysis Report Summary


About the Target
Based on the provided context information, the main viewpoints regarding MAPK3/ERK1 can be summarized as follows:
In the context of brain cancer cells, a proposed mechanism suggests that compressive force activates the MEK1/ERK1 pathway through EGFR/Ras/Raf activation. This activation regulates the expression of migration-related genes, such as GDF15, facilitating cell migration. Additionally, GDF15 may bind to its receptor and suppress MEK1/ERK1 activation to regulate its levels in compressed cells. Differential effects of mechanical compression on aggressive and less aggressive cells allow for a possible tumor-suppressing pathway to hinder the compression-induced migratory effect [1].
ERK1 interacts with BRD4 and inhibits its kinase activity. This interaction occurs in vivo and in situ, and ERK1 directly binds to the N-terminal region of BRD4. The competitive binding between ERK1, BRD4, and MYC suggests a complex regulatory relationship. Additionally, ERK1 phosphorylates MYC, leading to its degradation or stabilization, depending on the specific site of phosphorylation. This interplay contributes to the regulation of gene transcription [2].
In gastric cancer cells, the activation of M3 receptors by extracellular acetylcholine (ACh) leads to the activation of the EGFR/ERK1/2 signaling pathway. This activation promotes gastric cancer cell proliferation and survival. Blocking the M3 receptor or EGFR can weaken the role of ACh in cell proliferation and enhance chemosensitivity. Combining M3R/EGFR signaling inhibitors with traditional therapies may offer a potential strategy for gastric cancer treatment [3].
During infection with Toxoplasma gondii, the immediate-early phase involves the activation of ERK1/2 via TLR signaling, leading to the rapid expression of Egr-1 in dendritic cells (DCs). This activation is associated with cell activation and migration. In the extended phase, sustained Egr-1 expression is prompted by intracellular T. gondii replication and p38 phosphorylation, resulting in the down-modulation of DC maturation. Inhibition of ERK1/2 signaling abolishes the migratory responses of T. gondii-challenged DCs [4].
In neutrophils, YopH, a virulence factor of Yersinia pestis, inhibits various antimicrobial functions, including ROS production and degranulation. The adaptor protein SKAP2 is critical for ROS production but not for degranulation. While many other signaling proteins downstream of GPCRs are dispensable for SKAP2-mediated antimicrobial functions, this adaptor protein appears to function through a distinct signaling pathway. It plays a role in blocking certain integrin- and GPCR-dependent antimicrobial responses in neutrophils [5].
In summary, MAPK3/ERK1 is involved in various cellular processes, including cell migration, gene transcription regulation, cell proliferation, survival, and antimicrobial functions in different contexts. Its activation and interplay with other proteins can have diverse effects depending on the specific signaling pathway and cellular environment.
Based on the provided context information, the extracellular signal-regulated kinase 1 and 2 (ERK1/2) play a key role in endocytic trafficking and oncogenic signaling in non-small-cell lung cancer (NSCLC) cells [6]. Activation of FCHSD2, downstream of ERK1/2, enhances clathrin-mediated endocytosis (CME) and the initiation of clathrin-coated pits. Additionally, FCHSD2 promotes the trafficking of receptor tyrosine kinases (RTKs) from early endosomes to recycling endosomes, while negatively regulating Rab7 activity and late endosome/multivesicular body maturation. These activities of FCHSD2 alter the downstream signaling of RTKs and contribute to increased flux through the early endocytic pathways. Loss of FCHSD2 leads to the accumulation of RTKs in late endosomes/lysosomes, elevated levels of activated ERK1/2 in the nucleus, and increased transcription and expression of c-Jun, EGFR, and MET [6].
FCHSD2 expression gradually decreases in higher grades of lung adenocarcinoma tumors, while Rab7 expression shows the opposite correlation [6]. Furthermore, high FCHSD2 expression is associated with better survival rates in lung cancer patients, particularly in lung adenocarcinoma patients, whereas high Rab7 expression is linked to poorer survival rates [6].
In a separate study, downregulation of EBV-miR-BART1, a microRNA produced by Epstein-Barr virus (EBV), led to decreased migration and invasion of EBV-positive nasopharyngeal carcinoma (NPC) cells [7]. Reduction of EBV-miR-BART1 resulted in increased protein expressions of PTEN and E-cadherin, and decreased expressions of p-Akt, p-FAK, p-ERK1/2, N-cadherin, and vimentin in the cells [7]. EBV-miR-BART1 directly targets the tumor suppressor PTEN, reducing its dosage and activating PTEN-dependent pathways including PI3K-Akt, FAK-p130Cas, and Shc-MAPK/ERK1/2 signaling. This ultimately leads to actin cytoskeleton rearrangement and epithelial-to-mesenchymal transition (EMT), which promotes NPC migration, invasion, and metastasis [7].
In summary, ERK1/2, through its downstream target FCHSD2, regulates endocytic trafficking and plays a multifaceted role in oncogenic signaling in NSCLC cells [6]. FCHSD2 expression is associated with better survival rates in lung cancer patients, while Rab7 expression is associated with poorer survival rates [6]. Additionally, EBV-miR-BART1, a microRNA produced by EBV, promotes NPC metastasis by targeting PTEN and activating PTEN-dependent signaling pathways, including MAPK/ERK1/2 [7].
Figure [1]
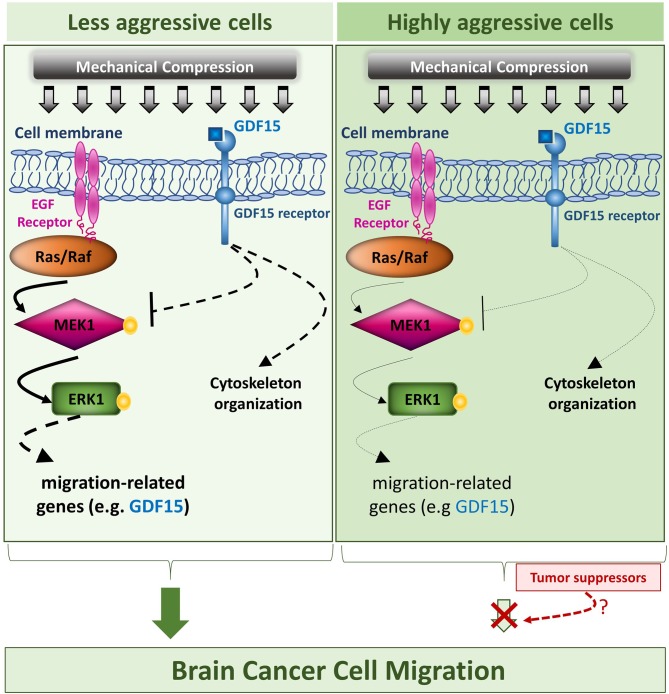
Figure [2]

Figure [3]
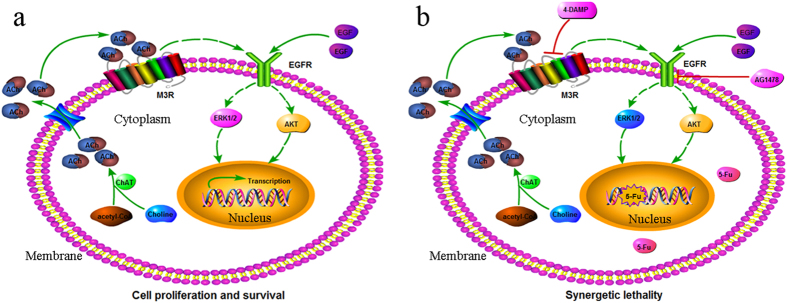
Figure [4]

Figure [5]
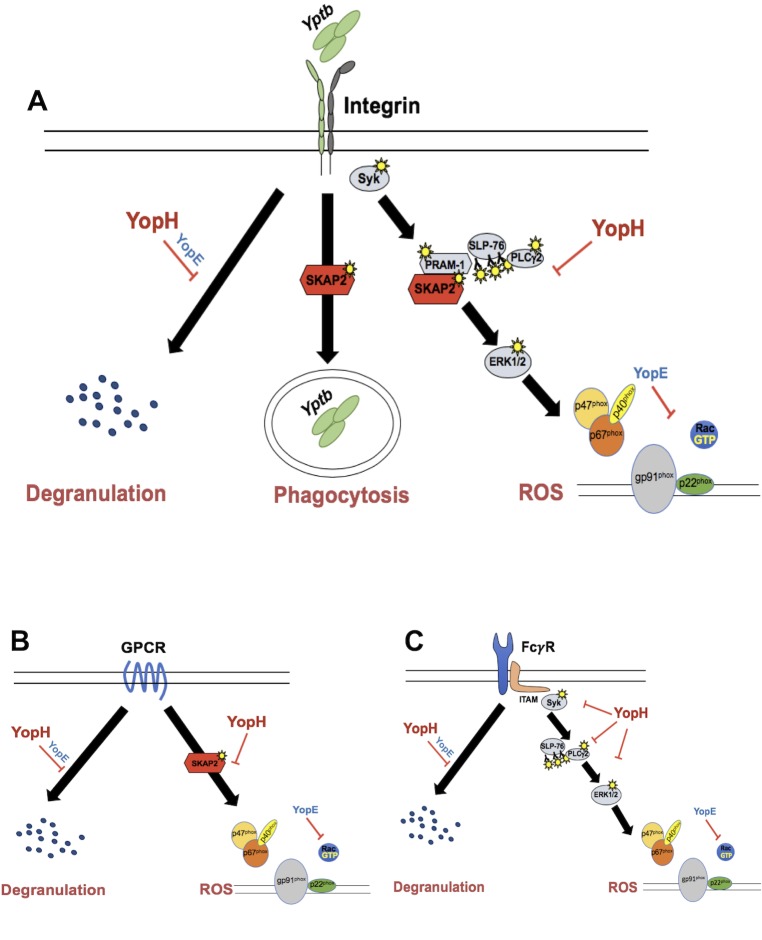
Figure [6]

Figure [7]

Note: If you are interested in the full version of this target analysis report, or if you'd like to learn how our AI-powered BDE-Chem can design therapeutic molecules to interact with the MAPK3 target at a cost 90% lower than traditional approaches, please feel free to contact us at BD@silexon.ai.
More Common Targets
ABCB1 | ABCG2 | ACE2 | AHR | AKT1 | ALK | AR | ATM | BAX | BCL2 | BCL2L1 | BECN1 | BRAF | BRCA1 | CAMP | CASP3 | CASP9 | CCL5 | CCND1 | CD274 | CD4 | CD8A | CDH1 | CDKN1A | CDKN2A | CREB1 | CXCL8 | CXCR4 | DNMT1 | EGF | EGFR | EP300 | ERBB2 | EREG | ESR1 | EZH2 | FN1 | FOXO3 | HDAC9 | HGF | HMGB1 | HSP90AA1 | HSPA4 | HSPA5 | IDO1 | IFNA1 | IGF1 | IGF1R | IL17A | IL6 | INS | JUN | KRAS | MAPK1 | MAPK14 | MAPK3 | MAPK8 | MAPT | MCL1 | MDM2 | MET | MMP9 | MTOR | MYC | NFE2L2 | NLRP3 | NOTCH1 | PARP1 | PCNA | PDCD1 | PLK1 | PRKAA1 | PRKAA2 | PTEN | PTGS2 | PTK2 | RELA | SIRT1 | SLTM | SMAD4 | SOD1 | SQSTM1 | SRC | STAT1 | STAT3 | STAT5A | TAK1 | TERT | TLR4 | TNF | TP53 | TXN | VEGFA | YAP1

